Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue
- PMID: 20238096
- PMCID: PMC2860566
- DOI: 10.1007/s00125-010-1701-4
Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue
Abstract
Aims/hypothesis: Increasing the expression of the brown adipose tissue-specific gene uncoupling protein-1 (Ucp1) is a potential target for treating obesity. We investigated the role of DNA methylation and histone modification in Ucp1 expression in adipose cell lines and ex vivo murine adipose tissues.
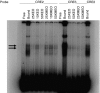
Methods: Methylation state of the Ucp1 enhancer was studied using bisulphite mapping in murine adipose cell lines, and tissue taken from cold-stressed mice, coupled with functional assays of the effects of methylation and demethylation of the Ucp1 promoter on gene expression and nuclear protein binding.
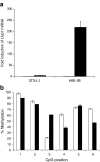
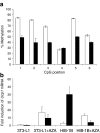
Results: We show that demethylation of the Ucp1 promoter by 5-aza-deoxycytidine increases Ucp1 expression while methylation of Ucp1 promoter-reporter constructs decreases expression. Brown adipose tissue-specific Ucp1 expression is associated with decreased CpG dinucleotide methylation of the Ucp1 enhancer. The lowest CpG dinucleotide methylation state was found in two cyclic AMP response elements (CRE3, CRE2) in the Ucp1 promoter and methylation of the CpG in CRE2, but not CRE3 decreased nuclear protein binding. Chromatin immunoprecipitation assays revealed the presence of the silencing DiMethH3K9 modification on the Ucp1 enhancer in white adipose tissue and the appearance of the active TriMethH3K4 mark at the Ucp1 promoter in brown adipose tissue in response to a cold environment.
Conclusions/interpretation: The results demonstrate that CpG dinucleotide methylation of the Ucp1 enhancer exhibits tissue-specific patterns in murine tissue and cell lines and suggest that adipose tissue-specific Ucp1 expression involves demethylation of CpG dinucleotides found in regulatory CREs in the Ucp1 enhancer, as well as modification of histone tails.
Figures


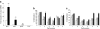
Similar articles
-
Orphan nuclear receptor NOR-1 enhances 3',5'-cyclic adenosine 5'-monophosphate-dependent uncoupling protein-1 gene transcription.Mol Endocrinol. 2008 May;22(5):1057-64. doi: 10.1210/me.2007-0464. Epub 2008 Jan 31. Mol Endocrinol. 2008. PMID: 18238829 Free PMC article.
-
UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic.Cell Rep. 2013 Dec 12;5(5):1196-203. doi: 10.1016/j.celrep.2013.10.044. Epub 2013 Nov 27. Cell Rep. 2013. PMID: 24290753
-
Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat.Mol Cell. 2015 Jan 22;57(2):235-46. doi: 10.1016/j.molcel.2014.12.005. Epub 2015 Jan 8. Mol Cell. 2015. PMID: 25578880 Free PMC article.
-
Transcriptional regulation of the uncoupling protein-1 gene.Biochimie. 2017 Mar;134:86-92. doi: 10.1016/j.biochi.2016.09.017. Epub 2016 Oct 5. Biochimie. 2017. PMID: 27693079 Review.
-
Is the heat surrounding adipose tissue mitochondria warranted?Curr Opin Clin Nutr Metab Care. 2014 Nov;17(6):503-8. doi: 10.1097/MCO.0000000000000102. Curr Opin Clin Nutr Metab Care. 2014. PMID: 25102333 Free PMC article. Review.
Cited by
-
Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity.Cell Metab. 2012 Dec 5;16(6):814-24. doi: 10.1016/j.cmet.2012.11.005. Cell Metab. 2012. PMID: 23217260 Free PMC article.
-
5-Aza-2'-Deoxycytidine Regulates White Adipocyte Browning by Modulating miRNA-133a/Prdm16.Metabolites. 2022 Nov 17;12(11):1131. doi: 10.3390/metabo12111131. Metabolites. 2022. PMID: 36422269 Free PMC article.
-
The Histone Demethylase UTX Promotes Brown Adipocyte Thermogenic Program Via Coordinated Regulation of H3K27 Demethylation and Acetylation.J Biol Chem. 2015 Oct 9;290(41):25151-63. doi: 10.1074/jbc.M115.662650. Epub 2015 Aug 25. J Biol Chem. 2015. PMID: 26306033 Free PMC article.
-
Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development.Biomolecules. 2021 Jun 7;11(6):851. doi: 10.3390/biom11060851. Biomolecules. 2021. PMID: 34200323 Free PMC article. Review.
-
Evidence of selection in the uncoupling protein 1 gene region suggests local adaptation to solar irradiance in savannah monkeys (Chlorocebus spp.).Proc Biol Sci. 2022 Sep 14;289(1982):20221254. doi: 10.1098/rspb.2022.1254. Epub 2022 Sep 14. Proc Biol Sci. 2022. PMID: 36100027 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

