Orchestrating serine resolvases
- PMID: 20298188
- PMCID: PMC3775659
- DOI: 10.1042/BST0380384
Orchestrating serine resolvases
Abstract
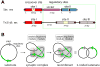
A remarkable feature of the serine resolvases is their regulation: the wild-type enzymes will catalyse intra- but not inter-molecular recombination, can sense the relative orientation of their sites and can exchange strands directionally, despite the fact that there is no net release of chemical bond energy. The key to this regulation is that they are only active within a large intertwined complex called the 'synaptosome'. Because substrate topology greatly facilitates (or, in other cases, inhibits) formation of the synaptosome, it acts as a 'topological filter'. Within the defined topology of the synaptosome, strand exchange releases supercoiling tension, providing an energy source to bias the reaction direction. The regulatory portion of this complex contains additional copies of the recombinase and sometimes other DNA-bending proteins. We are using a combination of X-ray crystallography, biochemistry and genetics to model the full synaptic complex and to understand how the regulatory portion activates the crossover-site-bound recombinases.
Figures



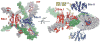
References
-
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of Site-Specific Recombination. Annu Rev Biochem. 2006;75:567–605. - PubMed
-
- Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley ND, Steitz TA. Structure of a synaptic gammadelta resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. - PubMed
-
- Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

