Engineering a family of synthetic splicing ribozymes
- PMID: 20299341
- PMCID: PMC2860135
- DOI: 10.1093/nar/gkq186
Engineering a family of synthetic splicing ribozymes
Abstract
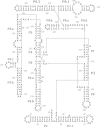
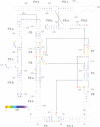
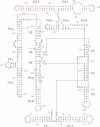
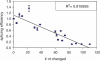
Controlling RNA splicing opens up possibilities for the synthetic biologist. The Tetrahymena ribozyme is a model group I self-splicing ribozyme that has been shown to be useful in synthetic circuits. To create additional splicing ribozymes that can function in synthetic circuits, we generated synthetic ribozyme variants by rationally mutating the Tetrahymena ribozyme. We present an alignment visualization for the ribozyme termed as structure information diagram that is similar to a sequence logo but with alignment data mapped on to secondary structure information. Using the alignment data and known biochemical information about the Tetrahymena ribozyme, we designed synthetic ribozymes with different primary sequences without altering the secondary structure. One synthetic ribozyme with 110 nt mutated retained 12% splicing efficiency in vivo. The results indicate that our biochemical understanding of the ribozyme is accurate enough to engineer a family of active splicing ribozymes with similar secondary structure but different primary sequences.
Figures
Similar articles
-
RNA reprogramming of alpha-mannosidase mRNA sequences in vitro by myxomycete group IC1 and IE ribozymes.FEBS J. 2006 Jun;273(12):2789-800. doi: 10.1111/j.1742-4658.2006.05295.x. FEBS J. 2006. PMID: 16817905
-
Imaging Tetrahymena ribozyme splicing activity in single live mammalian cells.Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):14892-6. doi: 10.1073/pnas.2036553100. Epub 2003 Nov 25. Proc Natl Acad Sci U S A. 2003. PMID: 14645710 Free PMC article.
-
Modulating the splicing activity of Tetrahymena ribozyme via RNA self-assembly.FEBS Lett. 2006 Mar 6;580(6):1592-6. doi: 10.1016/j.febslet.2006.01.090. Epub 2006 Feb 3. FEBS Lett. 2006. PMID: 16472807
-
Visualizing RNA splicing in vivo.Mol Biosyst. 2007 May;3(5):301-7. doi: 10.1039/b617574k. Epub 2007 Feb 21. Mol Biosyst. 2007. PMID: 17460789 Review.
-
Biological and functional aspects of catalytic RNAs.Crit Rev Eukaryot Gene Expr. 1992;2(4):331-57. Crit Rev Eukaryot Gene Expr. 1992. PMID: 1486242 Review.
Cited by
-
Roles of long-range tertiary interactions in limiting dynamics of the Tetrahymena group I ribozyme.J Am Chem Soc. 2014 May 7;136(18):6643-8. doi: 10.1021/ja413033d. Epub 2014 Apr 28. J Am Chem Soc. 2014. PMID: 24738560 Free PMC article.
-
Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of aGroup I Intron from Structured RNAs.Biology (Basel). 2016 Nov 17;5(4):43. doi: 10.3390/biology5040043. Biology (Basel). 2016. PMID: 27869660 Free PMC article.
-
Computational design of synthetic regulatory networks from a genetic library to characterize the designability of dynamical behaviors.Nucleic Acids Res. 2011 Nov 1;39(20):e138. doi: 10.1093/nar/gkr616. Epub 2011 Aug 24. Nucleic Acids Res. 2011. PMID: 21865275 Free PMC article.
-
A role for a single-stranded junction in RNA binding and specificity by the Tetrahymena group I ribozyme.J Am Chem Soc. 2012 Feb 1;134(4):1910-3. doi: 10.1021/ja2083575. Epub 2012 Jan 17. J Am Chem Soc. 2012. PMID: 22220837 Free PMC article.
References
-
- Che AJ. Engineering RNA logic with synthetic splicing ribozymes PhD thesis. Massachusetts Institute of Technology; 2008.
-
- Schultes EA, Bartel DP. One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science. 2000;289:448–452. - PubMed
-
- Cech TR. Self-splicing of group I introns. Ann. Rev. Biochem. 1990;59:543–568. - PubMed
-
- Woodson SA. Structure and assembly of group I introns. Curr. Opin. Struct. Biol. 2005;15:324–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

