Heritable individual-specific and allele-specific chromatin signatures in humans
- PMID: 20299549
- PMCID: PMC2929018
- DOI: 10.1126/science.1184655
Heritable individual-specific and allele-specific chromatin signatures in humans
Abstract
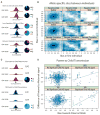
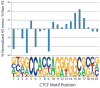
The extent to which variation in chromatin structure and transcription factor binding may influence gene expression, and thus underlie or contribute to variation in phenotype, is unknown. To address this question, we cataloged both individual-to-individual variation and differences between homologous chromosomes within the same individual (allele-specific variation) in chromatin structure and transcription factor binding in lymphoblastoid cells derived from individuals of geographically diverse ancestry. Ten percent of active chromatin sites were individual-specific; a similar proportion were allele-specific. Both individual-specific and allele-specific sites were commonly transmitted from parent to child, which suggests that they are heritable features of the human genome. Our study shows that heritable chromatin status and transcription factor binding differ as a result of genetic variation and may underlie phenotypic variation in humans.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

