Chromatin structure and the inheritance of epigenetic information
- PMID: 20300089
- PMCID: PMC3760772
- DOI: 10.1038/nrg2752
Chromatin structure and the inheritance of epigenetic information
Abstract
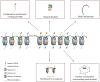
Although it is widely accepted that the regulation of the chromatin landscape is pivotal to conveying the epigenetic program, it is still unclear how a defined chromatin domain is reproduced following DNA replication and transmitted from one cell generation to the next. Here, we review the multiple mechanisms that potentially affect the inheritance of epigenetic information in somatic cells. We consider models of how histones might be recycled following replication, and discuss the importance of positive-feedback loops, long-range gene interactions and the complex network of trans-acting factors in the transmission of chromatin states.
Figures
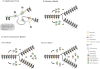


References
-
- Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. - PubMed
-
- Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–7. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

