Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence
- PMID: 20300646
- PMCID: PMC2837391
- DOI: 10.1371/journal.pone.0009688
Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence
Abstract
Background: Dinoflagellates are unicellular, often photosynthetic protists that play a major role in the dynamics of the Earth's oceans and climate. Sequencing of dinoflagellate nuclear DNA is thwarted by their massive genome sizes that are often several times that in humans. However, modern transcriptomic methods offer promising approaches to tackle this challenging system. Here, we used massively parallel signature sequencing (MPSS) to understand global transcriptional regulation patterns in Alexandrium tamarense cultures that were grown under four different conditions.
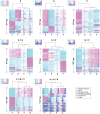
Methodology/principal findings: We generated more than 40,000 unique short expression signatures gathered from the four conditions. Of these, about 11,000 signatures did not display detectable differential expression patterns. At a p-value < 1E-10, 1,124 signatures were differentially expressed in the three treatments, xenic, nitrogen-limited, and phosphorus-limited, compared to the nutrient-replete control, with the presence of bacteria explaining the largest set of these differentially expressed signatures.
Conclusions/significance: Among microbial eukaryotes, dinoflagellates contain the largest number of genes in their nuclear genomes. These genes occur in complex families, many of which have evolved via recent gene duplication events. Our expression data suggest that about 73% of the Alexandrium transcriptome shows no significant change in gene expression under the experimental conditions used here and may comprise a "core" component for this species. We report a fundamental shift in expression patterns in response to the presence of bacteria, highlighting the impact of biotic interaction on gene expression in dinoflagellates.
Conflict of interest statement
Figures

References
-
- Jeong HJ, Latz MI. Growth and grazing rates of the heterotrophic dinoflagellates Protoperidinium spp. on red tide dinoflagellates. Mar Ecol Prog Ser. 1994;106:173–185.
-
- Drebes G, Schnepf E. Phagotrophy and development of Paulsenella cf chaetoceratis (Dinophyta), an ectoparasite of the diatom Streptotheca-thamesis. Helgol Meeresunters. 1982;35:501–515.
-
- Jacobson DM, Anderson DM. Widespread phagocytosis of ciliates and other protists by marine mixotrophic and heterotrophic thecate dinoflagellates. J Phycol. 1996;32:279–285.
-
- Jeong HJ, Du Yoo Y, Park JY, Song JY, Kim ST, et al. Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be mixotrophic. Aquat Microb Ecol. 2005;40:133–150.
-
- Pfiester LA, Anderson DM. Dinoflagellate reproduction. In: Taylor FJR, editor. The Biology of Dinoflagellates: Blackwell Science Inc; 1987. pp. 611–648.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

