Bias and evolution of the mutationally accessible phenotypic space in a developmental system
- PMID: 20300655
- PMCID: PMC2837400
- DOI: 10.1371/journal.pgen.1000877
Bias and evolution of the mutationally accessible phenotypic space in a developmental system
Abstract
Genetic and developmental architecture may bias the mutationally available phenotypic spectrum. Although such asymmetries in the introduction of variation may influence possible evolutionary trajectories, we lack quantitative characterization of biases in mutationally inducible phenotypic variation, their genotype-dependence, and their underlying molecular and developmental causes. Here we quantify the mutationally accessible phenotypic spectrum of the vulval developmental system using mutation accumulation (MA) lines derived from four wild isolates of the nematodes Caenorhabditis elegans and C. briggsae. The results confirm that on average, spontaneous mutations degrade developmental precision, with MA lines showing a low, yet consistently increased, proportion of developmental defects and variants. This result indicates strong purifying selection acting to maintain an invariant vulval phenotype. Both developmental system and genotype significantly bias the spectrum of mutationally inducible phenotypic variants. First, irrespective of genotype, there is a developmental bias, such that certain phenotypic variants are commonly induced by MA, while others are very rarely or never induced. Second, we found that both the degree and spectrum of mutationally accessible phenotypic variation are genotype-dependent. Overall, C. briggsae MA lines exhibited a two-fold higher decline in precision than the C. elegans MA lines. Moreover, the propensity to generate specific developmental variants depended on the genetic background. We show that such genotype-specific developmental biases are likely due to cryptic quantitative variation in activities of underlying molecular cascades. This analysis allowed us to identify the mutationally most sensitive elements of the vulval developmental system, which may indicate axes of potential evolutionary variation. Consistent with this scenario, we found that evolutionary trends in the vulval system concern the phenotypic characters that are most easily affected by mutation. This study provides an empirical assessment of developmental bias and the evolution of mutationally accessible phenotypes and supports the notion that such bias may influence the directions of evolutionary change.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


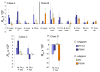
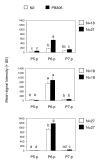
Similar articles
-
Cryptic genetic variation uncovers evolution of environmentally sensitive parameters in Caenorhabditis vulval development.Evol Dev. 2014 Sep;16(5):278-91. doi: 10.1111/ede.12091. Epub 2014 Aug 20. Evol Dev. 2014. PMID: 25143152
-
Role of pleiotropy in the evolution of a cryptic developmental variation in Caenorhabditis elegans.PLoS Biol. 2012 Jan;10(1):e1001230. doi: 10.1371/journal.pbio.1001230. Epub 2012 Jan 3. PLoS Biol. 2012. PMID: 22235190 Free PMC article.
-
The other side of phenotypic plasticity: a developmental system that generates an invariant phenotype despite environmental variation.J Biosci. 2009 Oct;34(4):543-51. doi: 10.1007/s12038-009-0073-8. J Biosci. 2009. PMID: 19920340 Review.
-
Genetic control of vulval development in Caenorhabditis briggsae.G3 (Bethesda). 2012 Dec;2(12):1625-41. doi: 10.1534/g3.112.004598. Epub 2012 Dec 1. G3 (Bethesda). 2012. PMID: 23275885 Free PMC article.
-
Robustness and flexibility in nematode vulva development.Trends Genet. 2012 Apr;28(4):185-95. doi: 10.1016/j.tig.2012.01.002. Epub 2012 Feb 9. Trends Genet. 2012. PMID: 22325232 Review.
Cited by
-
Induced mutations alter patterns of quantitative variation, phenotypic integration, and plasticity to elevated CO2 in Arabidopsis thaliana.J Plant Res. 2019 Jan;132(1):33-47. doi: 10.1007/s10265-018-1064-3. Epub 2018 Sep 25. J Plant Res. 2019. PMID: 30255212
-
The ancestral levels of transcription and the evolution of sexual phenotypes in filamentous fungi.PLoS Genet. 2017 Jul 13;13(7):e1006867. doi: 10.1371/journal.pgen.1006867. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28704372 Free PMC article.
-
Unmatched Level of Molecular Convergence among Deeply Divergent Complex Multicellular Fungi.Mol Biol Evol. 2020 Aug 1;37(8):2228-2240. doi: 10.1093/molbev/msaa077. Mol Biol Evol. 2020. PMID: 32191325 Free PMC article.
-
A larger target leads to faster evolution.Elife. 2020 Oct 14;9:e62689. doi: 10.7554/eLife.62689. Elife. 2020. PMID: 33052100 Free PMC article.
-
Evolution of New cis-Regulatory Motifs Required for Cell-Specific Gene Expression in Caenorhabditis.PLoS Genet. 2016 Sep 2;12(9):e1006278. doi: 10.1371/journal.pgen.1006278. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27588814 Free PMC article.
References
-
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genetics. 2007;8:803–813. - PubMed
-
- Lande R. Maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet Res. 1980;26:221–235. - PubMed
-
- Steppan SJ, Phillips PC, Houle D. Comparative quantitative genetics: evolution of the G matrix. TREE. 2002;17:320–327.
-
- Estes S, Phillips PC. Variation in pleiotropy and the mutational underpinnings of the G-matrix. Evolution. 2006;60:2655–2660. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

