Photocaged t7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells
- PMID: 20301166
- PMCID: PMC3762680
- DOI: 10.1002/cbic.201000041
Photocaged t7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells
Abstract
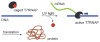
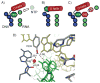
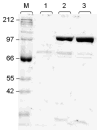
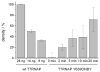
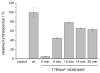
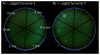
A light-activatable bacteriophage T7 RNA polymerase (T7RNAP) has been generated through the site-specific introduction of a photocaged tyrosine residue at the crucial position Tyr639 within the active site of the enzyme. The photocaged tyrosine disrupts polymerase activity by blocking the incoming nucleotide from reaching the active site of the enzyme. However, a brief irradiation with nonphototoxic UV light of 365 nm removes the ortho-nitrobenzyl caging group from Tyr639 and restores the RNA polymerase activity of T7RNAP. The complete orthogonality of T7RNAP to all endogenous RNA polymerases in pro- and eukaryotic systems allowed for the photochemical activation of gene expression in bacterial and mammalian cells. Specifically, E. coli cells were engineered to produce photocaged T7RNAP in the presence of a GFP reporter gene under the control of a T7 promoter. UV irradiation of these cells led to the spatiotemporal activation of GFP expression. In an analogous fashion, caged T7RNAP was transfected into human embryonic kidney (HEK293T) cells. Irradiation with UV light led to the activation of T7RNAP, thereby inducing RNA polymerization and expression of a luciferase reporter gene in tissue culture. The ability to achieve spatiotemporal regulation of orthogonal RNA synthesis enables the precise dissection and manipulation of a wide range of cellular events, including gene function.
Figures
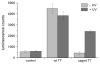
References
-
- Cramer P. BioEssays. 2002;24:724–729. - PubMed
-
- Deiters A. Chem Bio Chem. 2010;11:47–53. - PMC - PubMed
- Deiters A. Curr Opin Chem Biol. 2009;13:678–686. - PMC - PubMed
- Lee HM, Larson DR, Lawrence DS. ACS Chem Biol. 2009;4:409–427. - PMC - PubMed
- Casey JP, Blidner RA, Monroe WT. Mol Pharm. 2009;6:669–685. - PubMed
- Young DD, Deiters A. Org Biomol Chem. 2007;5:999–1005. - PubMed
- Tang X, Dmochowski IJ. Mol Biosyst. 2007;3:100–110. - PubMed
- Mayer G, Heckel A. Angew Chem. 2006;118:5020–5042.
- Angew Chem Int Ed. 2006;45:4900–4921. - PubMed
-
- Edwards WF, Young DD, Deiters A. ACS Chem Biol. 2009;4:441–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

