Identification of JAK-STAT pathways as important for the anti-inflammatory activity of a Hypericum perforatum fraction and bioactive constituents in RAW 264.7 mouse macrophages
- PMID: 20303133
- PMCID: PMC2858624
- DOI: 10.1016/j.phytochem.2010.02.006
Identification of JAK-STAT pathways as important for the anti-inflammatory activity of a Hypericum perforatum fraction and bioactive constituents in RAW 264.7 mouse macrophages
Abstract
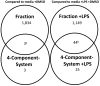
Hypericum perforatum extracts have been used to treat diseases, including mild-to-moderate depression and inflammatory conditions. It is particularly important to identify which constituents present in the H. perforatum extracts are responsible for its anti-inflammatory activity since consumers are taking H. perforatum preparations to treat inflammation. We used a combination of four putative bioactive constituents, called the 4-component-system that interacted synergistically to explain the light-activated anti-inflammatory activity of an H. perforatum fraction in RAW 264.7 mouse macrophages. We also combined the constituents at concentrations detected in the fraction to identify key molecular targets. LPS was used to model an inflammatory response, and the 4-component-system and H. perforatum fraction were used as treatments that inhibited LPS-induced prostaglandin E(2) (PGE(2)) production in RAW 264.7 mouse macrophages in the studies of gene expression profiles. We used Affymetrix genechips, statistical analysis, and quantitative real-time PCR to identify key gene targets of the 4-component-system and the sub-fraction from an H. perforatum ethanol extract. The H. perforatum sub-fraction, with or without LPS stimulation, affected far more genes than the 4-component-system with and without LPS. Genes involved in Janus kinase, as well as a signal transducer and activator of transcription (JAK-STAT) and eicosanoid pathways were identified that could account for the reduction in PGE(2) observed with both treatments in LPS-stimulated macrophages. Ten genes may be particularly important targets for activity of the 4-component-system and the fraction with LPS stimulation and these genes were involved in inflammatory signaling pathways, namely the JAK-STAT and eicosanoid pathways.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures



References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57:289–300.
-
- Bilia AR, Gallori S, Vincieri FF. St. John's wort and depression: Efficacy, safety and tolerability-an update. Life Sciences. 2002;70:3077–3096. - PubMed
-
- Bjorkbacka H, Fitzgerald KA, Huet F, Li X, Gregory JA, Lee MA, Ordija CM, Dowley NE, Golenbock DT, Freeman MW. The induction of macrophage gene expression by LPS predominantly utilizes MyD88-independent signaling cascades. Physiological Genomics. 2004;19:319–330. - PubMed
-
- Butterweck V, Lieflander-Wulf U, Winterhoff H, Nahrstedt A. Plasma levels of hypericin in presence of procyanidin B2 hyperoside: a pharmacokinetic study in rats. Planta Med. 2003;69:189–192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

