Bipedal locomotion in crawling cells
- PMID: 20303850
- PMCID: PMC2849068
- DOI: 10.1016/j.bpj.2009.10.058
Bipedal locomotion in crawling cells
Abstract
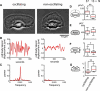
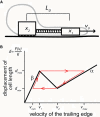
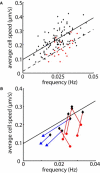
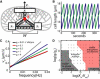
Many complex cellular processes from mitosis to cell motility depend on the ability of the cytoskeleton to generate force. Force-generating systems that act on elastic cytoskeletal elements are prone to oscillating instabilities. In this work, we have measured spontaneous shape and movement oscillations in motile fish epithelial keratocytes. In persistently polarized, fan-shaped cells, retraction of the trailing edge on one side of the cell body is out of phase with retraction on the other side, resulting in periodic lateral oscillation of the cell body. We present a physical description of keratocyte oscillation in which periodic retraction of the trailing edge is the result of elastic coupling with the leading edge. Consistent with the predictions of this model, the observed frequency of oscillation correlates with cell speed. In addition, decreasing the strength of adhesion to the substrate reduces the elastic force required for retraction, causing cells to oscillate with higher frequency at relatively lower speeds. These results demonstrate that simple elastic coupling between movement at the front of the cell and movement at the rear can generate large-scale mechanical integration of cell behavior.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Lauffenburger D.A., Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. - PubMed
-
- Keren K., Theriot J. Biophysical aspects of actin-based cell motility in fish epithelial keratocytes. In: Lenz P., editor. Cell Motility. Springer; New York: 2008. pp. 31–58.
-
- Gupton S.L., Waterman-Storer C.M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. - PubMed
-
- Theriot J.A., Mitchison T.J. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. - PubMed
-
- Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

