Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function
- PMID: 20304457
- PMCID: PMC3776475
- DOI: 10.1016/j.virol.2010.02.019
Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function
Erratum in
- Virology. 2010 Oct 10;406(1):162-3
Abstract
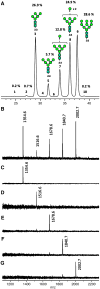
The HIV-1 envelope glycoprotein complex (Env) is the focus of vaccine development aimed at eliciting humoral immunity. Env's extensive and heterogeneous N-linked glycosylation affects folding, binding to lectin receptors, antigenicity and immunogenicity. We characterized recombinant Env proteins and virus particles produced in mammalian cells that lack N-acetylglucosaminyltransferase I (GnTI), an enzyme necessary for the conversion of oligomannose N-glycans to complex N-glycans. Carbohydrate analyses revealed that trimeric Env produced in GnTI(-/-) cells contained exclusively oligomannose N-glycans, with incompletely trimmed oligomannose glycans predominating. The folding and conformation of Env proteins was little affected by the manipulation of the glycosylation. Viruses produced in GnTI(-/-) cells were infectious, indicating that the conversion to complex glycans is not necessary for Env entry function, although virus binding to the C-type lectin DC-SIGN was enhanced. Manipulating Env's N-glycosylation may be useful for structural and functional studies and for vaccine design.
2010 Elsevier Inc. All rights reserved.
Figures




References
-
- Allaway GP, Davis-Bruno KL, Beaudry GA, Garcia EB, Wong EL, Ryder AM, Hasel KW, Gauduin MC, Koup RA, McDougal JS, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retroviruses. 1995;11(5):533–9. - PubMed
-
- Beddows S, Kirschner M, Campbell-Gardener L, Franti M, Dey AK, Iyer SP, Maddon PJ, Paluch M, Master A, Overbaugh J, VanCott T, Olson WC, Moore JP. Construction and characterization of soluble, cleaved, and stabilized trimeric Env proteins based on HIV type 1 Env subtype A. AIDS Res Hum Retroviruses. 2006;22(6):569–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

