The rules of DNA recognition by the androgen receptor
- PMID: 20304998
- PMCID: PMC5417492
- DOI: 10.1210/me.2009-0310
The rules of DNA recognition by the androgen receptor
Abstract
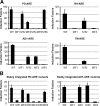
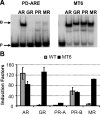
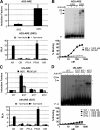
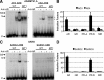
The androgen receptor (AR) and glucocorticoid, progestagen, and mineralocorticoid receptors all recognize classical DNA response elements that are organized as inverted repeats of 5'-AGAACA-3'-like motifs with a three-nucleotide spacer. Next to such elements, the AR also recognizes a second type of androgen response element (ARE), the so-called selective AREs, which resemble more the direct repeats of the same hexamer. In this work, we show that not only the AR but also the progestagen receptor can recognize the selective AREs, whereas neither glucocorticoid nor mineralocorticoid receptor can. Recently, genomic AR-binding fragments have been postulated to contain AR-binding sites that diverge considerably from the classical ARE consensus. Extensive mutational analyses of these candidate motifs, however, reinstalls the values of the consensus sequence for the AREs as mentioned above, the importance of their dimeric nature and the presence of exactly three-nucleotide spacing. We developed a position-specific probability matrix that was used to predict with higher accuracy new AREs in different AR-binding regions. So far, all AR-binding genomic fragments that were analyzed contain AREs defined as receptor-dimer binding motifs with the ability to confer responsiveness to a reporter gene.
Figures





Similar articles
-
Comparative analysis of the influence of the high-mobility group box 1 protein on DNA binding and transcriptional activation by the androgen, glucocorticoid, progesterone and mineralocorticoid receptors.Biochem J. 2002 Jan 1;361(Pt 1):97-103. doi: 10.1042/0264-6021:3610097. Biochem J. 2002. PMID: 11742533 Free PMC article.
-
Androgen-receptor-specific DNA binding to an element in the first exon of the human secretory component gene.Biochem J. 2001 Feb 1;353(Pt 3):611-20. doi: 10.1042/0264-6021:3530611. Biochem J. 2001. PMID: 11171058 Free PMC article.
-
Identification and characterization of androgen response elements.Methods Mol Biol. 2011;776:81-93. doi: 10.1007/978-1-61779-243-4_6. Methods Mol Biol. 2011. PMID: 21796522
-
Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation.J Steroid Biochem Mol Biol. 2001 Jan-Mar;76(1-5):23-30. doi: 10.1016/s0960-0760(00)00154-0. J Steroid Biochem Mol Biol. 2001. PMID: 11384860 Review.
-
Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression.Mol Genet Metab. 2003 Mar;78(3):175-85. doi: 10.1016/s1096-7192(03)00003-9. Mol Genet Metab. 2003. PMID: 12649062 Review.
Cited by
-
What Does Androgen Receptor Signaling Pathway in Sertoli Cells During Normal Spermatogenesis Tell Us?Front Endocrinol (Lausanne). 2022 Feb 24;13:838858. doi: 10.3389/fendo.2022.838858. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35282467 Free PMC article. Review.
-
Emerging mechanisms of enzalutamide resistance in prostate cancer.Nat Rev Urol. 2014 Dec;11(12):712-6. doi: 10.1038/nrurol.2014.243. Epub 2014 Sep 16. Nat Rev Urol. 2014. PMID: 25224448
-
Androgen receptor and miR-206 regulation in prostate cancer.Transcription. 2017;8(5):313-327. doi: 10.1080/21541264.2017.1322668. Epub 2017 Jun 9. Transcription. 2017. PMID: 28598253 Free PMC article. Review.
-
Stress, Sex, and Sugar: Glucocorticoids and Sex-Steroid Crosstalk in the Sex-Specific Misprogramming of Metabolism.J Endocr Soc. 2020 Jul 3;4(8):bvaa087. doi: 10.1210/jendso/bvaa087. eCollection 2020 Aug 1. J Endocr Soc. 2020. PMID: 32734132 Free PMC article. Review.
-
Targeting alternative sites on the androgen receptor to treat castration-resistant prostate cancer.Int J Mol Sci. 2013 Jun 14;14(6):12496-519. doi: 10.3390/ijms140612496. Int J Mol Sci. 2013. PMID: 23771019 Free PMC article. Review.
References
-
- Forman BM, Samuels HH1990. Interactions among a subfamily of nuclear hormone receptors: the regulatory zipper model. Mol Endocrinol 4:1293–1301 - PubMed
-
- Glass CK1994. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev 15:391–407 - PubMed
-
- Claessens F, Alen P, Devos A, Peeters B, Verhoeven G, Rombauts W1996. The androgen-specific probasin response element 2 interacts differentially with androgen and glucocorticoid receptors. J Biol Chem 271:19013–19016 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

