Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange
- PMID: 20305653
- PMCID: PMC2920209
- DOI: 10.1038/nsmb.1776
Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange
Abstract
DNA synthesis by classical polymerases can be blocked by many lesions. These blocks are overcome by translesion synthesis, whereby the stalled classical, replicative polymerase is replaced by a nonclassical polymerase. In eukaryotes this polymerase exchange requires proliferating cell nuclear antigen (PCNA) monoubiquitination. To better understand the polymerase exchange, we developed a means of producing monoubiquitinated PCNA, by splitting the protein into two self-assembling polypeptides. We determined the X-ray crystal structure of monoubiquitinated PCNA and found that the ubiquitin moieties are located on the back face of PCNA and interact with it through their canonical hydrophobic surface. Moreover, the attachment of ubiquitin does not change PCNA's conformation. We propose that PCNA ubiquitination facilitates nonclassical polymerase recruitment to the back of PCNA by forming a new binding surface for nonclassical polymerases, consistent with a 'tool belt' model of the polymerase exchange.
Figures
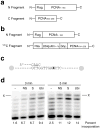
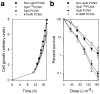

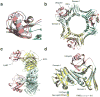
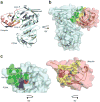

References
-
- Prakash S, Sung P, Prakash L. DNA-Repair Genes and Proteins of Saccharomyces-Cerevisiae. Annual Review of Genetics. 1993;27:33–70. - PubMed
-
- Jentsch S, McGrath JP, Varshavsky A. The Yeast DNA-Repair Gene Rad6 Encodes a Ubiquitin-Conjugating Enzyme. Nature. 1987;329:131–134. - PubMed
-
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific Complex-Formation between Yeast Rad6 and Rad18 Proteins - a Potential Mechanism for Targeting Rad6 Ubiquitin-Conjugating Activity to DNA-Damage Sites. Genes & Development. 1994;8:811–820. - PubMed
-
- Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. Journal of Biological Chemistry. 1997;272:23360–23365. - PubMed
-
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked tomodification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

