Conformational dynamics of single pre-mRNA molecules during in vitro splicing
- PMID: 20305654
- PMCID: PMC2881217
- DOI: 10.1038/nsmb.1767
Conformational dynamics of single pre-mRNA molecules during in vitro splicing
Abstract
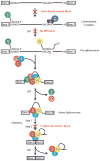
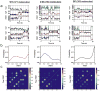
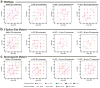
The spliceosome is a complex small nuclear RNA (snRNA)-protein machine that removes introns from pre-mRNAs via two successive phosphoryl transfer reactions. The chemical steps are isoenergetic, yet splicing requires at least eight RNA-dependent ATPases responsible for substantial conformational rearrangements. To comprehensively monitor pre-mRNA conformational dynamics, we developed a strategy for single-molecule FRET (smFRET) that uses a small, efficiently spliced yeast pre-mRNA, Ubc4, in which donor and acceptor fluorophores are placed in the exons adjacent to the 5' and 3' splice sites. During splicing in vitro, we observed a multitude of generally reversible time- and ATP-dependent conformational transitions of individual pre-mRNAs. The conformational dynamics of branchpoint and 3'-splice site mutants differ from one another and from wild type. Because all transitions are reversible, spliceosome assembly appears to be occurring close to thermal equilibrium.
Figures






References
-
- Jurica MS, Moore MJ. Pre–mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. - PubMed
-
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–26. - PubMed
-
- Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA–dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–91. - PubMed
-
- Couto JR, Tamm J, Parker R, Guthrie C. A trans–acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1:445–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

