A three-stage genome-wide association study of general cognitive ability: hunting the small effects
- PMID: 20306291
- PMCID: PMC2992848
- DOI: 10.1007/s10519-010-9350-4
A three-stage genome-wide association study of general cognitive ability: hunting the small effects
Abstract
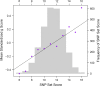
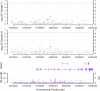
Childhood general cognitive ability (g) is important for a wide range of outcomes in later life, from school achievement to occupational success and life expectancy. Large-scale association studies will be essential in the quest to identify variants that make up the substantial genetic component implicated by quantitative genetic studies. We conducted a three-stage genome-wide association study for general cognitive ability using over 350,000 single nucleotide polymorphisms (SNPs) in the quantitative extremes of a population sample of 7,900 7-year-old children from the UK Twins Early Development Study. Using two DNA pooling stages to enrich true positives, each of around 1,000 children selected from the extremes of the distribution, and a third individual genotyping stage of over 3,000 children to test for quantitative associations across the normal range, we aimed to home in on genes of small effect. Genome-wide results suggested that our approach was successful in enriching true associations and 28 SNPs were taken forward to individual genotyping in an unselected population sample. However, although we found an enrichment of low P values and identified nine SNPs nominally associated with g (P < 0.05) that show interesting characteristics for follow-up, further replication will be necessary to meet rigorous standards of association. These replications may take advantage of SNP sets to overcome limitations of statistical power. Despite our large sample size and three-stage design, the genes associated with childhood g remain tantalizingly beyond our current reach, providing further evidence for the small effect sizes of individual loci. Larger samples, denser arrays and multiple replications will be necessary in the hunt for the genetic variants that influence human cognitive ability.
Figures


References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

