Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua)
- PMID: 20307277
- PMCID: PMC2846918
- DOI: 10.1186/1471-2164-11-191
Development of a SNP resource and a genetic linkage map for Atlantic cod (Gadus morhua)
Abstract
Background: Atlantic cod (Gadus morhua) is a species with increasing economic significance for the aquaculture industry. The genetic improvement of cod will play a critical role in achieving successful large-scale aquaculture. While many microsatellite markers have been developed in cod, the number of single nucleotide polymorphisms (SNPs) is currently limited. Here we report the identification of SNPs from sequence data generated by a large-scale expressed sequence tag (EST) program, focusing on fish originating from Canadian waters.
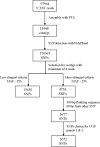
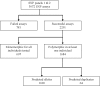
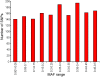
Results: A total of 97976 ESTs were assembled to generate 13448 contigs. We detected 4753 SNPs that met our selection criteria (depth of coverage > or = 4 reads; minor allele frequency > 25%). 3072 SNPs were selected for testing. The percentage of successful assays was 75%, with 2291 SNPs amplifying correctly. Of these, 607 (26%) SNPs were monomorphic for all populations tested. In total, 64 (4%) of SNPs are likely to represent duplicated genes or highly similar members of gene families, rather than alternative alleles of the same gene, since they showed a high frequency of heterozygosity. The remaining polymorphic SNPs (1620) were categorised as validated SNPs. The mean minor allele frequency of the validated loci was 0.258 (+/- 0.141). Of the 1514 contigs from which validated SNPs were selected, 31% have a significant blast hit. For the SNPs predicted to occur in coding regions (141), we determined that 36% (51) are non-synonymous. Many loci (1033 SNPs; 64%) are polymorphic in all populations tested. However a small number of SNPs (184) that are polymorphic in the Western Atlantic were monomorphic in fish tested from three European populations. A preliminary linkage map has been constructed with 23 major linkage groups and 924 mapped SNPs.
Conclusions: These SNPs represent powerful tools to accelerate the genetic improvement of cod aquaculture. They have been used to build a genetic linkage map that can be applied to quantitative trait locus (QTL) discovery. Since these SNPs were generated from ESTs, they are linked to specific genes. Genes that map within QTL intervals can be prioritized for testing to determine whether they contribute to observed phenotypes.
Figures




References
-
- Rose GA. Cod: The ecological history of the North Atlantic fisheries. Breakwater Books, St. John's, Newfoundland; 2007.
-
- Rosenlund G, Skretting M. Worldwide status and perspective on gadoid aquaculture. ICES Journal of Marine Science. 2006;63:194–197. doi: 10.1016/j.icesjms.2005.11.012. - DOI
-
- Brown JA, Minkoff G, Puvanendran V. Larviculture of Atlantic cod (Gadus morhua): progress, protocols and problems. Aquaculture. 2003;227:357–372. doi: 10.1016/S0044-8486(03)00514-3. - DOI
-
- Gjedrem T. Genetic improvement of coldwater fish species. Aquaculture Research. 2000;31:25–33. doi: 10.1046/j.1365-2109.2000.00389.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

