Molecular basis for the high degree of antigenic cross-reactivity between hepatitis B virus capsids (HBcAg) and dimeric capsid-related protein (HBeAg): insights into the enigmatic nature of the e-antigen
- PMID: 20307545
- PMCID: PMC2860019
- DOI: 10.1016/j.jmb.2010.03.026
Molecular basis for the high degree of antigenic cross-reactivity between hepatitis B virus capsids (HBcAg) and dimeric capsid-related protein (HBeAg): insights into the enigmatic nature of the e-antigen
Abstract
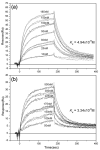
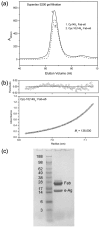
The hepatitis B virus core gene codes for two closely related antigens: a 21-kDa protein that forms dimers that assemble as multimegadalton capsids, and a 17-kDa protein that also forms dimers but that do not assemble. The proteins, respectively referred to as core antigen (HBcAg) and e-antigen (HBeAg), share a sequence of 149 residues but have different amino- and carboxy-termini. Their structural and serological relationship has long been unclear. With insights gained from recent structural studies on immune complexes of the capsids, the relationship was reassessed using recombinant forms of the antigens and a panel of monoclonal antibodies (mAbs) commonly believed to discriminate between core and e-antigen. Surface plasmon resonance (SPR) was used to measure the affinities, in contrast to previous studies that used more error-prone and less sensitive plate-type assays. Four of the six mAbs did not discriminate between core and e-antigen, nor did they discriminate between e-antigen and dimers of dissociated core antigen capsids. One mAb (3120) was specific for assembled capsids and one (e6) was specific for unassembled dimers. Epitope valency of the e-antigen was also studied, using a sandwich SPR assay where e-antigen was captured with one mAb and probed with a second. The e-antigen is often considered to be a monomeric protein on the basis of monovalent reactivity with antibody pairs specific for either an alpha or beta epitope (in a prior nomenclature for e-antigen specificity). This model, however, is incorrect, because recombinant e-antigen is a stable dimer and its apparent monovalency is due to steric blockage. This was proven by the formation of a 2:1 Fab e6-e-antigen complex. These results suggest new approaches for the isolation of the authentic e-antigen, its biological assay, and its stabilization as an immune complex for structural studies.
(c) 2010 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Specific determination of hepatitis B e antigen by antibodies targeting precore unique epitope facilitates clinical diagnosis and drug evaluation against hepatitis B virus infection.Emerg Microbes Infect. 2021 Dec;10(1):37-50. doi: 10.1080/22221751.2020.1862631. Emerg Microbes Infect. 2021. PMID: 33296295 Free PMC article.
-
Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens.J Mol Biol. 1998 Jun 26;279(5):1111-21. doi: 10.1006/jmbi.1998.1845. J Mol Biol. 1998. PMID: 9642088
-
Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly.Biochemistry. 1995 Apr 18;34(15):4919-32. doi: 10.1021/bi00015a003. Biochemistry. 1995. PMID: 7711014
-
The hepatitis B virus core and e antigens elicit different Th cell subsets: antigen structure can affect Th cell phenotype.J Virol. 1997 Mar;71(3):2192-201. doi: 10.1128/JVI.71.3.2192-2201.1997. J Virol. 1997. PMID: 9032353 Free PMC article.
-
Development of hepatitis B virus capsids into a whole-chain protein antigen display platform: new particulate Lyme disease vaccines.Int J Med Microbiol. 2008 Jan;298(1-2):135-42. doi: 10.1016/j.ijmm.2007.08.002. Epub 2007 Sep 20. Int J Med Microbiol. 2008. PMID: 17888729 Review.
Cited by
-
Core protein: A pleiotropic keystone in the HBV lifecycle.Antiviral Res. 2015 Sep;121:82-93. doi: 10.1016/j.antiviral.2015.06.020. Epub 2015 Jun 27. Antiviral Res. 2015. PMID: 26129969 Free PMC article. Review.
-
Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.PLoS Comput Biol. 2020 Apr 20;16(4):e1007782. doi: 10.1371/journal.pcbi.1007782. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32310951 Free PMC article.
-
HBV Core Protein Is in Flux between Cytoplasmic, Nuclear, and Nucleolar Compartments.mBio. 2021 Feb 9;12(1):e03514-20. doi: 10.1128/mBio.03514-20. mBio. 2021. PMID: 33563815 Free PMC article.
-
Hepatitis B Virus Nucleocapsid Assembly.J Mol Biol. 2025 Apr 30:169182. doi: 10.1016/j.jmb.2025.169182. Online ahead of print. J Mol Biol. 2025. PMID: 40316009 Review.
-
Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.Int J Mol Sci. 2021 Aug 24;22(17):9140. doi: 10.3390/ijms22179140. Int J Mol Sci. 2021. PMID: 34502049 Free PMC article.
References
-
- Lesmana LA, Leung NW, Mahachai V, Phiet PH, Suh DJ, Yao G, Zhuang H. Hepatitis B: overview of the burden of disease in the Asia-Pacific region. Liver Int. 2006;26 2:3–10. - PubMed
-
- Liaw YF. Management of chronic hepatitis B: an evolving issue. Liver Int. 2006;26 2:1–2. - PubMed
-
- Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE. Fields Virology. 3. Lippincott-Raven Publishers; Philadelphia: 1996.
-
- Ou JH. Molecular biology of hepatitis B virus e antigen. J Gastroenterol Hepatol. 1997;12:S178–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

