Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing
- PMID: 20307546
- PMCID: PMC2878758
- DOI: 10.1016/j.jmb.2010.03.025
Structure-guided mutational analysis of a yeast DEAD-box protein involved in mitochondrial RNA splicing
Abstract
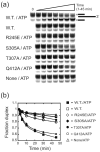
DEAD-box proteins are RNA-dependent ATPase enzymes that have been implicated in nearly all aspects of RNA metabolism. Since many of these enzymes have been shown to possess common biochemical properties in vitro, including the ability to bind and hydrolyze ATP, to bind nucleic acid, and to promote helix unwinding, DEAD-box proteins are generally thought to modulate RNA structure in vivo. However, the extent to which these enzymatic properties are important for the in vivo functions of DEAD-box proteins remains unclear. To evaluate how these properties influence DEAD-box protein native function, we probed the importance of several highly conserved residues in the yeast DEAD-box protein Mss116p, which is required for the splicing of all mitochondrial catalytic introns in Saccharomyces cerevisiae. Using an MSS116 deletion strain, we have expressed plasmid-borne variants of MSS116 containing substitutions in residues predicted to be important for extensive networks of interactions required for ATP hydrolysis and helix unwinding. We have analyzed the importance of these residues to the splicing functions of Mss116p in vivo and compared these results with the biochemical properties of recombinant proteins determined here and in previously published work. We observed that the efficiency by which an Mss116p variant catalyzes ATP hydrolysis correlates with facilitating mitochondrial splicing, while efficient helix unwinding appears to be insufficient for splicing. In addition, we show that each splicing-defective variant affects the splicing of structurally diverse introns to the same degree. Together, these observations suggest that the efficiency by which Mss116p catalyzes the hydrolysis of ATP is critical for all of its splicing functions in vivo. Given that ATP hydrolysis stimulates the recycling of DEAD-box proteins, these observations support a model in which enzyme turnover is a crucial factor in Mss116p splicing function. These results are discussed in the context of current models of Mss116p-facilitated splicing.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures



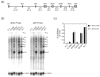
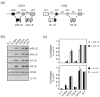
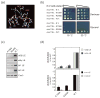
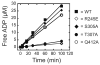
Similar articles
-
ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo.J Mol Biol. 2011 Aug 19;411(3):661-79. doi: 10.1016/j.jmb.2011.05.047. Epub 2011 Jun 7. J Mol Biol. 2011. PMID: 21679717 Free PMC article.
-
Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity.J Mol Biol. 2007 Jan 19;365(3):835-55. doi: 10.1016/j.jmb.2006.09.083. Epub 2006 Oct 3. J Mol Biol. 2007. PMID: 17081564 Free PMC article.
-
Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro.J Mol Biol. 2008 Feb 1;375(5):1344-64. doi: 10.1016/j.jmb.2007.11.041. Epub 2007 Nov 22. J Mol Biol. 2008. PMID: 18096186 Free PMC article.
-
Mss116p: a DEAD-box protein facilitates RNA folding.RNA Biol. 2013 Jan;10(1):71-82. doi: 10.4161/rna.22492. Epub 2012 Oct 12. RNA Biol. 2013. PMID: 23064153 Free PMC article. Review.
-
Helicases involved in splicing from malaria parasite Plasmodium falciparum.Parasitol Int. 2011 Dec;60(4):335-40. doi: 10.1016/j.parint.2011.09.007. Epub 2011 Oct 1. Parasitol Int. 2011. PMID: 21996352 Review.
Cited by
-
Single-molecule analysis of Mss116-mediated group II intron folding.Nature. 2010 Oct 21;467(7318):935-9. doi: 10.1038/nature09422. Epub 2010 Oct 13. Nature. 2010. PMID: 20944626 Free PMC article.
-
Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins.J Mol Biol. 2011 Jun 10;409(3):399-414. doi: 10.1016/j.jmb.2011.04.004. Epub 2011 Apr 9. J Mol Biol. 2011. PMID: 21501623 Free PMC article.
-
High-throughput genetic identification of functionally important regions of the yeast DEAD-box protein Mss116p.J Mol Biol. 2011 Nov 11;413(5):952-72. doi: 10.1016/j.jmb.2011.09.015. Epub 2011 Sep 16. J Mol Biol. 2011. PMID: 21945532 Free PMC article.
-
ATP utilization and RNA conformational rearrangement by DEAD-box proteins.Annu Rev Biophys. 2012;41:247-67. doi: 10.1146/annurev-biophys-050511-102243. Epub 2012 Feb 13. Annu Rev Biophys. 2012. PMID: 22404686 Free PMC article. Review.
-
RNA helicases and remodeling proteins.Curr Opin Chem Biol. 2011 Oct;15(5):636-42. doi: 10.1016/j.cbpa.2011.07.019. Epub 2011 Aug 20. Curr Opin Chem Biol. 2011. PMID: 21862383 Free PMC article. Review.
References
-
- Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem 2009 - PubMed
-
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. - PubMed
-
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

