LVIS553 transcriptional regulator specifically recognizes novobiocin as an effector molecule
- PMID: 20308066
- PMCID: PMC2878039
- DOI: 10.1074/jbc.M110.111138
LVIS553 transcriptional regulator specifically recognizes novobiocin as an effector molecule
Abstract
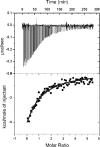
In this study we aimed to identify small molecules with high affinity involved in the allosteric regulation of LVIS553, a MarR member from Lactobacillus brevis ATCC367. Using high throughput screening, novobiocin was found to specifically bind LVIS553 with a K(D) = 33.8 +/- 2.9 microM consistent with a biologically relevant ligand. Structure guided site-directed mutagenesis identified Lys(9) as a key residue in novobiocin recognition. The results found in vitro were correlated in vivo. An increased tolerance to the antibiotic was observed when LVIS553 and the downstream putative transport protein LVIS552 were either expressed in a low copy plasmid in L. brevis or as a single copy chromosomal insertion in Bacillus subtilis. We provide evidence that LVIS553 is involved in the specific regulation of a new mechanism of tolerance to novobiocin.
Figures




Similar articles
-
Lactobacillus brevis responds to flavonoids through KaeR, a LysR-type of transcriptional regulator.Mol Microbiol. 2011 Sep;81(6):1623-39. doi: 10.1111/j.1365-2958.2011.07796.x. Epub 2011 Aug 22. Mol Microbiol. 2011. PMID: 21819457
-
A dual role of the transcriptional regulator TstR provides insights into cyanide detoxification in Lactobacillus brevis.Mol Microbiol. 2014 May;92(4):853-71. doi: 10.1111/mmi.12598. Epub 2014 Apr 14. Mol Microbiol. 2014. PMID: 24684290 Free PMC article.
-
Mutations That Enhance the Ciprofloxacin Resistance of Escherichia coli with qnrA1.Antimicrob Agents Chemother. 2015 Dec 28;60(3):1537-45. doi: 10.1128/AAC.02167-15. Antimicrob Agents Chemother. 2015. PMID: 26711751 Free PMC article.
-
Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins.Curr Issues Mol Biol. 2006 Jan;8(1):51-62. Curr Issues Mol Biol. 2006. PMID: 16450885 Review.
-
MarR family transcription factors: dynamic variations on a common scaffold.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):595-613. doi: 10.1080/10409238.2017.1344612. Epub 2017 Jul 3. Crit Rev Biochem Mol Biol. 2017. PMID: 28670937 Review.
Cited by
-
The transcriptional activator LdtR from 'Candidatus Liberibacter asiaticus' mediates osmotic stress tolerance.PLoS Pathog. 2014 Apr 24;10(4):e1004101. doi: 10.1371/journal.ppat.1004101. eCollection 2014 Apr. PLoS Pathog. 2014. PMID: 24763829 Free PMC article.
-
The Sdp-SH3b2 domain contained in Lactobacillus johnsonii N6.2-derived extracellular vesicles inhibit murine norovirus replication.Front Immunol. 2024 Dec 5;15:1490755. doi: 10.3389/fimmu.2024.1490755. eCollection 2024. Front Immunol. 2024. PMID: 39712028 Free PMC article.
-
A serralysin-like protein of Candidatus Liberibacter asiaticus modulates components of the bacterial extracellular matrix.Front Microbiol. 2022 Oct 19;13:1006962. doi: 10.3389/fmicb.2022.1006962. eCollection 2022. Front Microbiol. 2022. PMID: 36338045 Free PMC article.
-
Anaerobic p-coumarate degradation by Rhodopseudomonas palustris and identification of CouR, a MarR repressor protein that binds p-coumaroyl coenzyme A.J Bacteriol. 2012 Apr;194(8):1960-7. doi: 10.1128/JB.06817-11. Epub 2012 Feb 10. J Bacteriol. 2012. PMID: 22328668 Free PMC article.
-
Identification of a Ligand Binding Pocket in LdtR from Liberibacter asiaticus.Front Microbiol. 2015 Nov 25;6:1314. doi: 10.3389/fmicb.2015.01314. eCollection 2015. Front Microbiol. 2015. PMID: 26635775 Free PMC article.
References
-
- Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. (2001) Nat. Struct. Biol. 8, 710–714 - PubMed
-
- Saridakis V., Shahinas D., Xu X., Christendat D. (2008) J. Mol. Biol. 377, 655–667 - PubMed
-
- Alekshun M. N., Kim Y. S., Levy S. B. (2000) Mol. Microbiol. 35, 1394–1404 - PubMed
-
- Okada N., Oi Y., Takeda-Shitaka M., Kanou K., Umeyama H., Haneda T., Miki T., Hosoya S., Danbara H. (2007) Microbiology 153, 548–560 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

