Topologically random insertion of EmrE supports a pathway for evolution of inverted repeats in ion-coupled transporters
- PMID: 20308069
- PMCID: PMC2865334
- DOI: 10.1074/jbc.M110.108746
Topologically random insertion of EmrE supports a pathway for evolution of inverted repeats in ion-coupled transporters
Abstract
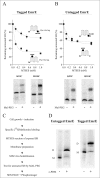
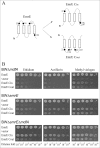
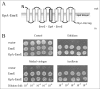
Inverted repeats in ion-coupled transporters have evolved independently in many unrelated families. It has been suggested that this inverted symmetry is an essential element of the mechanism that allows for the conformational transitions in transporters. We show here that small multidrug transporters offer a model for the evolution of such repeats. This family includes both homodimers and closely related heterodimers. In the former, the topology determinants, evidently identical in each protomer, are weak, and we show that for EmrE, an homodimer from Escherichia coli, the insertion into the membrane is random, and dimers are functional whether they insert into the cytoplasmic membrane with the N- and C-terminal domains facing the inside or the outside of the cell. Also, mutants designed to insert with biased topology are functional regardless of the topology. In the case of EbrAB, a heterodimer homologue supposed to interact antiparallel, we show that one of the subunits, EbrB, can also function as a homodimer, most likely in a parallel mode. In addition, the EmrE homodimer can be forced to an antiparallel topology by fusion of an additional transmembrane segment. The simplicity of the mechanism of coupling ion and substrate transport and the few requirements for substrate recognition provide the robustness necessary to tolerate such a unique and unprecedented ambiguity in the interaction of the subunits and in the dimer topology relative to the membrane. The results suggest that the small multidrug transporters are at an evolutionary junction and provide a model for the evolution of structure of transport proteins.
Figures



Similar articles
-
Inducing conformational preference of the membrane protein transporter EmrE through conservative mutations.Elife. 2019 Oct 22;8:e48909. doi: 10.7554/eLife.48909. Elife. 2019. PMID: 31637997 Free PMC article.
-
Emulating membrane protein evolution by rational design.Science. 2007 Mar 2;315(5816):1282-4. doi: 10.1126/science.1135406. Epub 2007 Jan 25. Science. 2007. PMID: 17255477
-
Parallel topology of genetically fused EmrE homodimers.EMBO J. 2008 Jan 9;27(1):17-26. doi: 10.1038/sj.emboj.7601951. Epub 2007 Dec 6. EMBO J. 2008. PMID: 18059473 Free PMC article.
-
EmrE, a model for studying evolution and mechanism of ion-coupled transporters.Biochim Biophys Acta. 2009 May;1794(5):748-62. doi: 10.1016/j.bbapap.2008.12.018. Epub 2009 Jan 3. Biochim Biophys Acta. 2009. PMID: 19167526 Review.
-
Precious things come in little packages.J Mol Microbiol Biotechnol. 2001 Apr;3(2):155-62. J Mol Microbiol Biotechnol. 2001. PMID: 11321568 Review.
Cited by
-
Comparison of the functional properties of trimeric and monomeric CaiT of Escherichia coli.Sci Rep. 2019 Mar 7;9(1):3787. doi: 10.1038/s41598-019-40516-7. Sci Rep. 2019. PMID: 30846799 Free PMC article.
-
Transforming a drug/H+ antiporter into a polyamine importer by a single mutation.Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16894-9. doi: 10.1073/pnas.1211831109. Epub 2012 Oct 3. Proc Natl Acad Sci U S A. 2012. PMID: 23035252 Free PMC article.
-
Exploring Uniform, Dual, and Dynamic Topologies of Membrane Proteins by Substituted Cysteine Accessibility Method (SCAM™).Methods Mol Biol. 2024;2715:121-157. doi: 10.1007/978-1-0716-3445-5_9. Methods Mol Biol. 2024. PMID: 37930526 Free PMC article.
-
New substrates on the block: clinically relevant resistances for EmrE and homologues.J Bacteriol. 2012 Dec;194(24):6766-70. doi: 10.1128/JB.01318-12. Epub 2012 Oct 5. J Bacteriol. 2012. PMID: 23042996 Free PMC article.
-
Proof of dual-topology architecture of Fluc F- channels with monobody blockers.Nat Commun. 2014 Oct 7;5:5120. doi: 10.1038/ncomms6120. Nat Commun. 2014. PMID: 25290819 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

