Transposable elements in natural populations of Drosophila melanogaster
- PMID: 20308097
- PMCID: PMC2871824
- DOI: 10.1098/rstb.2009.0318
Transposable elements in natural populations of Drosophila melanogaster
Abstract
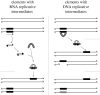
Transposable elements (TEs) are families of small DNA sequences found in the genomes of virtually all organisms. The sequences typically encode essential components for the replicative transposition sequences of that TE family. Thus, TEs are simply genomic parasites that inflict detrimental mutations on the fitness of their hosts. Several models have been proposed for the containment of TE copy number in outbreeding host populations such as Drosophila. Surveys of the TEs in genomes from natural populations of Drosophila have played a central role in the investigation of TE dynamics. The early surveys indicated that a typical TE insertion is rare in a population, which has been interpreted as evidence that each TE is selected against. The proposed mechanisms of this natural selection are reviewed here. Subsequent and more targeted surveys identify heterogeneity among types of TEs and also highlight the large role of homologous and possibly ectopic crossing over in the dynamics of the Drosophila TEs. The recent discovery of germline-specific RNA interference via the piwi-interacting RNA pathway opens yet another interesting mechanism that may be critical in containing the copy number of TEs in natural populations of Drosophila. The expected flood of Drosophila population genomics is expected to rapidly advance understanding of the dynamics of TEs.
Figures
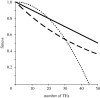
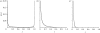
 . These three figures depict the impact of β as population size, Ne, goes from 104 (β = 0.4) to 105 (β = 4) and then to 106 (β = 40). The probability a TE insertion will occur at intermediate or high frequencies increases dramatically. Other assumed parameters are u = 10−5, T = 4 × 104 and
. These three figures depict the impact of β as population size, Ne, goes from 104 (β = 0.4) to 105 (β = 4) and then to 106 (β = 40). The probability a TE insertion will occur at intermediate or high frequencies increases dramatically. Other assumed parameters are u = 10−5, T = 4 × 104 and  . (a) α = 0.0005, β = 0.4; (b) α = 0.005, β = 4 and (c) α = 0.05, β = 40.
. (a) α = 0.0005, β = 0.4; (b) α = 0.005, β = 4 and (c) α = 0.05, β = 40.
References
-
- Aminetzach Y. T., Macpherson J. M., Petrov D. A.2005Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309, 764–767 (doi:10.1126/science.1112699) - DOI - PubMed
-
- Aravin A. A., Hannon G. J., Brennecke J.2007The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764 (doi:10.1126/science.1146484) - DOI - PubMed
-
- Bartolome C., Maside X.2004The lack of recombination drives the fixation of transposable elements on the fourth chromosome of Drosophila melanogaster. Genet. Res. 83, 91–100 (doi:10.1017/S0016672304006755) - DOI - PubMed
-
- Bartolome C., Maside X., Charlesworth B.2002On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19, 926–937 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

