Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction
- PMID: 20308164
- PMCID: PMC2910054
- DOI: 10.1093/nar/gkq174
Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction
Abstract
The human APOBEC3 proteins are DNA cytidine deaminases that impede the replication of many different transposons and viruses. The genes that encode APOBEC3A, APOBEC3B, APOBEC3C, APOBEC3D, APOBEC3F, APOBEC3G and APOBEC3H were generated through relatively recent recombination events. The resulting high degree of inter-relatedness has complicated the development of specific quantitative PCR assays for these genes despite considerable interest in understanding their expression profiles. Here, we describe a set of quantitative PCR assays that specifically measures the mRNA levels of each APOBEC3 gene. The specificity and sensitivity of each assay was validated using a full matrix of APOBEC3 cDNA templates. The assays were used to quantify the APOBEC3 repertoire in multiple human T-cell lines, bulk leukocytes and leukocyte subsets, and 20 different human tissues. The data demonstrate that multiple APOBEC3 genes are expressed constitutively in most types of cells and tissues, and that distinct APOBEC3 genes are induced upon T-cell activation and interferon treatment. These data help define the APOBEC3 repertoire relevant to HIV-1 restriction in T cells, and they suggest a general model in which multiple APOBEC3 proteins function together to provide a constitutive barrier to foreign genetic elements, which can be fortified by transcriptional induction.
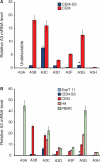
Figures





References
-
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. - PubMed
-
- Malim MH, Emerman M. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. - PubMed
-
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

