Identification and characterization of beta-lactamase inhibitor protein-II (BLIP-II) interactions with beta-lactamases using phage display
- PMID: 20308189
- PMCID: PMC2865362
- DOI: 10.1093/protein/gzq017
Identification and characterization of beta-lactamase inhibitor protein-II (BLIP-II) interactions with beta-lactamases using phage display
Abstract
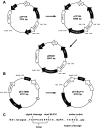
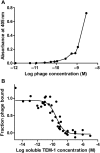
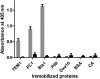
Protein-protein interactions are critical to cellular processes yet the ability to predict and rationally design interactions is limited because of incomplete knowledge of the principles governing these interactions. The beta-lactamase inhibitory protein (BLIP)/beta-lactamase interaction has become a model system to investigate protein-protein interactions and has been the focus of several structural, thermodynamic and binding specificity studies. BLIP-II also inhibits beta-lactamase but has no sequence homology with BLIP. The structure of BLIP-II in complex with TEM-1 beta-lactamase revealed that BLIP-II has a completely different structure than BLIP but it interacts with the same protruding loop-helix region of TEM-1 as does BLIP. The significance of the individual interacting residues in molecular recognition by BLIP-II is currently unknown. Therefore, a phage display vector was developed with the purpose of expressing BLIP-II onto the surface of the M13 filamentous bacteriophage. The BLIP-II displayed phage bound to TEM-1 with picomolar affinity indicating that BLIP-II is properly folded while on the surface of the phage. The phage system, as well as enzyme inhibition assays with purified proteins, revealed that BLIP-II is a more potent inhibitor than BLIP for several class A beta-lactamases with K(i) values in the low picomolar range.
Figures

Similar articles
-
Identification of residues in beta-lactamase critical for binding beta-lactamase inhibitory protein.J Biol Chem. 1999 Mar 12;274(11):6963-71. doi: 10.1074/jbc.274.11.6963. J Biol Chem. 1999. PMID: 10066750
-
Structural and computational characterization of the SHV-1 beta-lactamase-beta-lactamase inhibitor protein interface.J Biol Chem. 2006 Sep 8;281(36):26745-53. doi: 10.1074/jbc.M603878200. Epub 2006 Jun 29. J Biol Chem. 2006. PMID: 16809340
-
Identification of the β-lactamase inhibitor protein-II (BLIP-II) interface residues essential for binding affinity and specificity for class A β-lactamases.J Biol Chem. 2013 Jun 14;288(24):17156-66. doi: 10.1074/jbc.M113.463521. Epub 2013 Apr 27. J Biol Chem. 2013. PMID: 23625930 Free PMC article.
-
Insight into Structure-Function Relationships of β-Lactamase and BLIPs Interface Plasticity using Protein-Protein Interactions.Curr Pharm Des. 2019;25(31):3378-3389. doi: 10.2174/1381612825666190911154650. Curr Pharm Des. 2019. PMID: 31544712 Review.
-
Structure, Function of Serine and Metallo-β-lactamases and their Inhibitors.Curr Protein Pept Sci. 2018;19(2):130-144. doi: 10.2174/0929866524666170724160623. Curr Protein Pept Sci. 2018. PMID: 28745223 Review.
Cited by
-
Characterization of the global stabilizing substitution A77V and its role in the evolution of CTX-M β-lactamases.Antimicrob Agents Chemother. 2015 Nov;59(11):6741-8. doi: 10.1128/AAC.00618-15. Epub 2015 Aug 17. Antimicrob Agents Chemother. 2015. PMID: 26282414 Free PMC article.
-
A drug-resistant β-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency.J Biol Chem. 2020 Dec 25;295(52):18239-18255. doi: 10.1074/jbc.RA120.016103. Epub 2020 Oct 26. J Biol Chem. 2020. PMID: 33109613 Free PMC article.
-
Optimising CNT-FET biosensor design through modelling of biomolecular electrostatic gating and its application to β-lactamase detection.Nat Commun. 2024 Aug 29;15(1):7482. doi: 10.1038/s41467-024-51325-6. Nat Commun. 2024. PMID: 39209826 Free PMC article.
-
Systematic substitutions at BLIP position 50 result in changes in binding specificity for class A β-lactamases.BMC Biochem. 2017 Mar 6;18(1):2. doi: 10.1186/s12858-017-0077-1. BMC Biochem. 2017. PMID: 28264645 Free PMC article.
-
Enhancing the activity of β-lactamase inhibitory protein-II with cell-penetrating peptide against KPC-2-carrying Klebsiella pneumoniae.PLoS One. 2024 Jan 26;19(1):e0296727. doi: 10.1371/journal.pone.0296727. eCollection 2024. PLoS One. 2024. PMID: 38277388 Free PMC article.
References
-
- Adediran S.A., Zhang Z., Nukaga M., Palzkill T., Pratt R.F. Biochemistry. 2005;44:7543–7552. doi:10.1021/bi050136f. - DOI - PubMed
-
- Albeck S., Schreiber G. Biochemistry. 1999;38:11–21. doi:10.1021/bi981772z. - DOI - PubMed
-
- Alkhatib G. Curr. Opin. HIV AIDS. 2009;4:96–103. doi:10.1097/COH.0b013e328324bbec. - DOI - PMC - PubMed
-
- Bass S., Greene R., Wells J.A. Proteins. 1990;8:309–314. doi:10.1002/prot.340080405. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

