The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice
- PMID: 20308426
- PMCID: PMC2845082
- DOI: 10.1083/jcb.200909117
The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice
Abstract
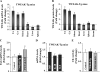
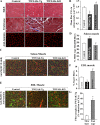
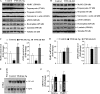
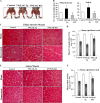
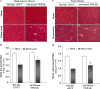
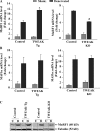
Skeletal muscle atrophy occurs in a variety of clinical settings, including cachexia, disuse, and denervation. Inflammatory cytokines have been shown to be mediators of cancer cachexia; however, the role of cytokines in denervation- and immobilization-induced skeletal muscle loss remains unknown. In this study, we demonstrate that a single cytokine, TNF-like weak inducer of apoptosis (TWEAK), mediates skeletal muscle atrophy that occurs under denervation conditions. Transgenic expression of TWEAK induces atrophy, fibrosis, fiber-type switching, and the degradation of muscle proteins. Importantly, genetic ablation of TWEAK decreases the loss of muscle proteins and spared fiber cross-sectional area, muscle mass, and strength after denervation. Expression of the TWEAK receptor Fn14 (fibroblast growth factor-inducible receptor 14) and not the cytokine is significantly increased in muscle upon denervation, demonstrating an unexpected inside-out signaling pathway; the receptor up-regulation allows for TWEAK activation of nuclear factor kappaB, causing an increase in the expression of the E3 ubiquitin ligase MuRF1. This study reveals a novel mediator of skeletal muscle atrophy and indicates that the TWEAK-Fn14 system is an important target for preventing skeletal muscle wasting.
Figures



References
-
- Acharyya S., Butchbach M.E., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., et al. 2005. Dystrophin glycoprotein complex dysfunction: a regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 8:421–432 10.1016/j.ccr.2005.10.004 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

