Short N-terminal sequences package proteins into bacterial microcompartments
- PMID: 20308536
- PMCID: PMC2867708
- DOI: 10.1073/pnas.0913199107
Short N-terminal sequences package proteins into bacterial microcompartments
Abstract
Hundreds of bacterial species produce proteinaceous microcompartments (MCPs) that act as simple organelles by confining the enzymes of metabolic pathways that have toxic or volatile intermediates. A fundamental unanswered question about bacterial MCPs is how enzymes are packaged within the protein shell that forms their outer surface. Here, we report that a short N-terminal peptide is necessary and sufficient for packaging enzymes into the lumen of an MCP involved in B(12)-dependent 1,2-propanediol utilization (Pdu MCP). Deletion of 10 or 14 amino acids from the N terminus of the propionaldehyde dehydrogenase (PduP) enzyme, which is normally found within the Pdu MCP, substantially impaired packaging, with minimal effects on its enzymatic activity. Fusion of the 18 N-terminal amino acids from PduP to GFP, GST, or maltose-binding protein resulted in their encapsulation within MCPs. Bioinformatic analyses revealed N-terminal extensions in two additional Pdu proteins and three proteins from two unrelated MCPs, suggesting that N-terminal peptides may be used to package proteins into diverse MCPs. The potential uses of MCP assembly principles in nature and in biotechnology are discussed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



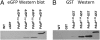


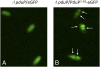

Comment in
-
Addressing microbial organelles: a short peptide directs enzymes to the interior.Proc Natl Acad Sci U S A. 2010 Apr 27;107(17):7627-8. doi: 10.1073/pnas.1003433107. Epub 2010 Apr 16. Proc Natl Acad Sci U S A. 2010. PMID: 20400692 Free PMC article. No abstract available.
References
-
- Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. - PubMed
-
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Microbiol. 2008;6:681–691. - PubMed
-
- Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

