Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances
- PMID: 20308540
- PMCID: PMC2851944
- DOI: 10.1073/pnas.1001945107
Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances
Abstract
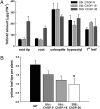
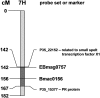
The aim of the present study was to assess possible adverse effects of transgene expression in leaves of field-grown barley relative to the influence of genetic background and the effect of plant interaction with arbuscular mycorrhizal fungi. We conducted transcript profiling, metabolome profiling, and metabolic fingerprinting of wild-type accessions and barley transgenics with seed-specific expression of (1,3-1, 4)-beta-glucanase (GluB) in Baronesse (B) as well as of transgenics in Golden Promise (GP) background with ubiquitous expression of codon-optimized Trichoderma harzianum endochitinase (ChGP). We found more than 1,600 differential transcripts between varieties GP and B, with defense genes being strongly overrepresented in B, indicating a divergent response to subclinical pathogen challenge in the field. In contrast, no statistically significant differences between ChGP and GP could be detected based on transcriptome or metabolome analysis, although 22 genes and 4 metabolites were differentially abundant when comparing GluB and B, leading to the distinction of these two genotypes in principle component analysis. The coregulation of most of these genes in GluB and GP, as well as simple sequence repeat-marker analysis, suggests that the distinctive alleles in GluB are inherited from GP. Thus, the effect of the two investigated transgenes on the global transcript profile is substantially lower than the effect of a minor number of alleles that differ as a consequence of crop breeding. Exposing roots to the spores of the mycorrhizal Glomus sp. had little effect on the leaf transcriptome, but central leaf metabolism was consistently altered in all genotypes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cellini F, et al. Unintended effects and their detection in genetically modified crops. Food Chem Toxicol. 2004;42:1089–1125. - PubMed
-
- Schäfer P, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica . Plant J. 2009;59:461–474. - PubMed
-
- Trethewey RN. Metabolite profiling as an aid to metabolic engineering in plants. Curr Opin Plant Biol. 2004;7:196–201. - PubMed
-
- Gregersen PL, Brinch-Pedersen H, Holm PB. A microarray-based comparative analysis of gene expression profiles during grain development in transgenic and wild type wheat. Transgenic Res. 2005;14:887–905. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

