Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21
- PMID: 20331851
- PMCID: PMC2853522
- DOI: 10.1186/1471-2164-11-195
Molecular and evolutionary characteristics of the fraction of human alpha satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21
Abstract
Background: The mode of evolution of the highly homogeneous Higher-Order-Repeat-containing alpha satellite arrays is still subject to discussion. This is also true of the CENP-A associated repeats where the centromere is formed.
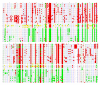
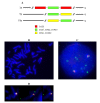
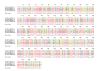
Results: In this paper, we show that the molecular mechanisms by which these arrays evolve are identical in multiple chromosomes: i) accumulation of crossovers that homogenise and expand the arrays into different domains and subdomains that are mostly unshared between homologues and ii) sporadic mutations and conversion events that simultaneously differentiate them from one another. Individual arrays are affected by these mechanisms to different extents that presumably increase with time. Repeats associated with CENP-A, where the centromere is formed, are subjected to the same evolutionary mechanisms, but constitute minor subsets that exhibit subtle sequence differences from those of the bulk repeats. While the DNA sequence per se is not essential for centromere localisation along an array, it appears that certain sequences can be selected against. On chromosomes 1 and 19, which are more affected by the above evolutionary mechanisms than are chromosomes 21 and 5, CENP-A associated repeats were also recovered from a second homogeneous array present on each chromosome. This could be a way for chromosomes to sustain mitosis and meiosis when the normal centromere locus is ineluctably undermined by the above mechanisms.
Conclusion: We discuss, in light of these observations, possible scenarios for the normal evolutionary fates of human centromeric regions.
Figures









Similar articles
-
Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C.Dev Cell. 2017 Aug 7;42(3):226-240.e6. doi: 10.1016/j.devcel.2017.07.001. Dev Cell. 2017. PMID: 28787590 Free PMC article.
-
Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells.Chromosome Res. 2011 May;19(4):457-70. doi: 10.1007/s10577-011-9208-5. Epub 2011 Apr 12. Chromosome Res. 2011. PMID: 21484447 Free PMC article.
-
Organization and molecular evolution of CENP-A--associated satellite DNA families in a basal primate genome.Genome Biol Evol. 2011;3:1136-49. doi: 10.1093/gbe/evr083. Epub 2011 Aug 9. Genome Biol Evol. 2011. PMID: 21828373 Free PMC article.
-
The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres.Chromosome Res. 2004;12(6):543-56. doi: 10.1023/B:CHRO.0000036593.72788.99. Chromosome Res. 2004. PMID: 15289662 Review.
-
Genetic and epigenetic regulation of centromeres: a look at HAC formation.Chromosome Res. 2015 Feb;23(1):87-103. doi: 10.1007/s10577-015-9470-z. Chromosome Res. 2015. PMID: 25682171 Review.
Cited by
-
Centromeres under Pressure: Evolutionary Innovation in Conflict with Conserved Function.Genes (Basel). 2020 Aug 10;11(8):912. doi: 10.3390/genes11080912. Genes (Basel). 2020. PMID: 32784998 Free PMC article. Review.
-
Centromere Repeats: Hidden Gems of the Genome.Genes (Basel). 2019 Mar 16;10(3):223. doi: 10.3390/genes10030223. Genes (Basel). 2019. PMID: 30884847 Free PMC article. Review.
-
RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin.Elife. 2017 Aug 1;6:e25299. doi: 10.7554/eLife.25299. Elife. 2017. PMID: 28760200 Free PMC article.
-
Alpha satellite DNA biology: finding function in the recesses of the genome.Chromosome Res. 2018 Sep;26(3):115-138. doi: 10.1007/s10577-018-9582-3. Epub 2018 Jul 5. Chromosome Res. 2018. PMID: 29974361 Free PMC article. Review.
-
Aneuploidy and DNA Methylation as Mirrored Features of Early Human Embryo Development.Genes (Basel). 2020 Sep 17;11(9):1084. doi: 10.3390/genes11091084. Genes (Basel). 2020. PMID: 32957536 Free PMC article. Review.
References
-
- Mahtani MM, Willard HF. Physical and genetic mapping of the human X chromosome centromere: repression of recombination. Genome Res. 1998;8:100–110. - PubMed
-
- Choo KHA. Why is the centromere so cold? Genome Res. 1998;8:81–82. - PubMed
-
- Puechberty J, Laurent AM, Gimenez S, Billault A, Brun-Laurent ME, Calenda A, Marcais B, Prades C, Ioannou P, Yurov Y, Roizès G. Genetic and physical analyses of the pericentromeric regions of human chromosomes 5 and 19. Recombination across 5cen. Genomics. 1999;56:274–287. doi: 10.1006/geno.1999.5742. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

