NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex
- PMID: 20332092
- PMCID: PMC2871465
- DOI: 10.1074/jbc.M110.117069
NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex
Abstract
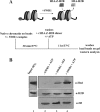
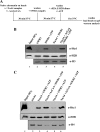
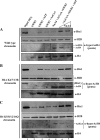
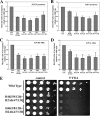
Structural and functional analyses of nucleosomes containing histone variant H2A.Z have drawn a lot of interest over the past few years. Important work in budding yeast has shown that H2A.Z (Htz1)-containing nucleosomes are specifically located on the promoter regions of genes, creating a specific chromatin structure that is poised for disassembly during transcription activation. The SWR1 complex is responsible for incorporation of Htz1 into nucleosomes through ATP-dependent exchange of canonical H2A-H2B dimers for Htz1-H2B dimers. Interestingly, the yeast SWR1 complex is functionally linked to the NuA4 acetyltransferase complex in vivo. NuA4 and SWR1 are physically associated in higher eukaryotes as they are homologous to the TIP60/p400 complex, which encompasses both histone acetyltransferase (Tip60) and histone exchange (p400/Domino) activities. Here we present work investigating the impact of NuA4-dependent acetylation on SWR1-driven incorporation of H2A.Z into chromatin. Using in vitro histone exchange assays with native chromatin, we demonstrate that prior chromatin acetylation by NuA4 greatly stimulates the exchange of H2A for H2A.Z. Interestingly, we find that acetylation of H2A or H4 N-terminal tails by NuA4 can independently stimulate SWR1 activity. Accordingly, we demonstrate that mutations of H4 or H2A N-terminal lysine residues have similar effects on H2A.Z incorporation in vivo, and cells carrying mutations in both tails are nonviable. Finally, depletion experiments indicate that the bromodomain-containing protein Bdf1 is important for NuA4-dependent stimulation of SWR1. These results provide important mechanistic insight into the functional cross-talk between chromatin acetylation and ATP-dependent exchange of histone H2A variants.
Figures



References
-
- Kouzarides T. (2007) Cell 128, 693–705 - PubMed
-
- Clapier C. R., Cairns B. R. (2009) Annu. Rev. Biochem. 78, 273–304 - PubMed
-
- Kamakaka R. T., Biggins S. (2005) Genes Dev. 19, 295–310 - PubMed
-
- Draker R., Cheung P. (2009) Biochem. Cell Biol. 87, 19–25 - PubMed
-
- Raisner R. M., Madhani H. D. (2006) Curr. Opin. Genet. Dev. 16, 119–124 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

