Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of beta-catenin mRNA
- PMID: 20332102
- PMCID: PMC2848118
- DOI: 10.1242/jcs.056176
Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of beta-catenin mRNA
Abstract
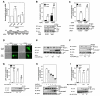
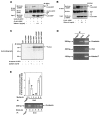
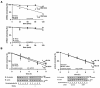
Canonical Wnt/beta-catenin signaling is crucial during embryonic development. Upon Wnt stimulation, Dishevelled proteins relay the signal from upstream Frizzled receptors to downstream effectors. By using affinity purification followed by ion-trap mass spectrometry we identified K-homology splicing regulator protein (KSRP) as a novel Dishevelled-interacting protein. We show that KSRP negatively regulates Wnt/beta-catenin signaling at the level of post-transcriptional CTNNB1 (beta-catenin) mRNA stability. Thus, Dishevelled-KSRP complex operates in Wnt regulation of beta-catenin, functioning post-transcriptionally upon CTNNB1 mRNA stability.
Figures




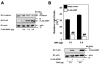

Similar articles
-
Dishevelled-2 docks and activates Src in a Wnt-dependent manner.J Cell Sci. 2009 Dec 15;122(Pt 24):4439-51. doi: 10.1242/jcs.051847. Epub 2009 Nov 17. J Cell Sci. 2009. PMID: 19920076 Free PMC article.
-
A novel Wilms tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway.J Biol Chem. 2010 May 7;285(19):14585-93. doi: 10.1074/jbc.M109.094334. Epub 2010 Mar 10. J Biol Chem. 2010. PMID: 20220130 Free PMC article.
-
Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl.Mol Cell. 2010 Mar 12;37(5):607-19. doi: 10.1016/j.molcel.2010.01.035. Mol Cell. 2010. PMID: 20227366
-
Dishevelled: The hub of Wnt signaling.Cell Signal. 2010 May;22(5):717-27. doi: 10.1016/j.cellsig.2009.11.021. Epub 2009 Dec 13. Cell Signal. 2010. PMID: 20006983 Review.
-
New steps in the Wnt/beta-catenin signal transduction pathway.Recent Prog Horm Res. 2000;55:225-36. Recent Prog Horm Res. 2000. PMID: 11036939 Review.
Cited by
-
KHSRP-bound small nucleolar RNAs associate with promotion of cell invasiveness and metastasis of pancreatic cancer.Oncotarget. 2020 Jan 14;11(2):131-147. doi: 10.18632/oncotarget.27413. eCollection 2020 Jan 14. Oncotarget. 2020. PMID: 32010427 Free PMC article.
-
miR-3619-3p promotes papillary thyroid carcinoma progression via Wnt/β-catenin pathway.Ann Transl Med. 2019 Nov;7(22):643. doi: 10.21037/atm.2019.10.71. Ann Transl Med. 2019. PMID: 31930044 Free PMC article.
-
KSRP-mediated Wnt/β-catenin activation promotes follicular thyroid cancer progression and stemness.Br J Cancer. 2025 Aug 23. doi: 10.1038/s41416-025-03142-x. Online ahead of print. Br J Cancer. 2025. PMID: 40849355
-
Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer.Nat Cell Biol. 2016 Aug;18(8):897-909. doi: 10.1038/ncb3380. Epub 2016 Jun 27. Nat Cell Biol. 2016. PMID: 27347849
-
The RNA-Binding Protein KSRP Modulates Cytokine Expression of CD4+ T Cells.J Immunol Res. 2019 Aug 14;2019:4726532. doi: 10.1155/2019/4726532. eCollection 2019. J Immunol Res. 2019. PMID: 31511826 Free PMC article.
References
-
- Aghib D. F., McCrea P. D. (1995). The E-cadherin complex contains the src substrate p120. Exp. Cell Res. 218, 359-369 - PubMed
-
- Behrens J., von Kries J. P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. (1996). Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382, 638-642 - PubMed
-
- Bikkavilli R. K., Feigin M. E., Malbon C. C. (2008). G alpha o mediates WNT-JNK signaling through dishevelled 1 and 3, RhoA family members, and MEKK 1 and 4 in mammalian cells. J. Cell Sci. 121, 234-245 - PubMed
-
- Boutros M., Paricio N., Strutt D. I., Mlodzik M. (1998). Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94, 109-118 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

