Similarly strong purifying selection acts on human disease genes of all evolutionary ages
- PMID: 20333184
- PMCID: PMC2817408
- DOI: 10.1093/gbe/evp013
Similarly strong purifying selection acts on human disease genes of all evolutionary ages
Abstract
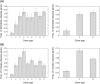
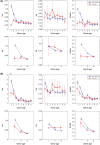
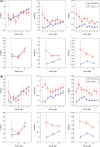
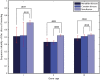
A number of studies have showed that recently created genes differ from the genes created in deep evolutionary past in many aspects. Here, we determined the age of emergence and propensity for gene loss (PGL) of all human protein-coding genes and compared disease genes with non-disease genes in terms of their evolutionary rate, strength of purifying selection, mRNA expression, and genetic redundancy. The older and the less prone to loss, non-disease genes have been evolving 1.5- to 3-fold slower between humans and chimps than young non-disease genes, whereas Mendelian disease genes have been evolving very slowly regardless of their ages and PGL. Complex disease genes showed an intermediate pattern. Disease genes also have higher mRNA expression heterogeneity across multiple tissues than non-disease genes regardless of age and PGL. Young and middle-aged disease genes have fewer similar paralogs as non-disease genes of the same age. We reasoned that genes were more likely to be involved in human disease if they were under a strong functional constraint, expressed heterogeneously across tissues, and lacked genetic redundancy. Young human genes that have been evolving under strong constraint between humans and chimps might also be enriched for genes that encode important primate or even human-specific functions.
Keywords: evolutionary age of genes; human disease genes; propensity for gene loss; strength of selection.
Figures

References
-
- Alba MM, Castresana J. Inverse relationship between evolutionary rate and age of mammalian genes. Mol Biol Evol. 2005;22:598–606. - PubMed
-
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

