A complex suite of forces drives gene traffic from Drosophila X chromosomes
- PMID: 20333188
- PMCID: PMC2817413
- DOI: 10.1093/gbe/evp018
A complex suite of forces drives gene traffic from Drosophila X chromosomes
Abstract
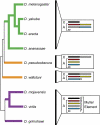
Theoretical studies predict X chromosomes and autosomes should be under different selection pressures, and there should therefore be differences in sex-specific and sexually antagonistic gene content between the X and the autosomes. Previous analyses have identified an excess of genes duplicated by retrotransposition from the X chromosome in Drosophila melanogaster. A number of hypotheses may explain this pattern, including mutational bias, escape from X-inactivation during spermatogenesis, and the movement of male-favored (sexually antagonistic) genes from a chromosome that is predominantly carried by females. To distinguish among these processes and to examine the generality of these patterns, we identified duplicated genes in nine sequenced Drosophila genomes. We find that, as in D. melanogaster, there is an excess of genes duplicated from the X chromosome across the genus Drosophila. This excess duplication is due almost completely to genes duplicated by retrotransposition, with little to no excess from the X among genes duplicated via DNA intermediates. The only exception to this pattern appears within the burst of duplication that followed the creation of the Drosophila pseudoobscura neo-X chromosome. Additionally, we examined genes relocated among chromosomal arms (i.e., genes duplicated to new locations coupled with the loss of the copy in the ancestral locus) and found an excess of genes relocated off the ancestral X and neo-X chromosomes. Interestingly, many of the same genes were duplicated or relocated from the independently derived neo-X chromosomes of D. pseudoobscura and Drosophila willistoni, suggesting that natural selection favors the traffic of genes from X chromosomes. Overall, we find that the forces driving gene duplication from X chromosomes are dependent on the lineage in question, the molecular mechanism of duplication considered, the preservation of the ancestral copy, and the age of the X chromosome.
Keywords: X-inactivation; gene duplication; neo-X chromosomes; retrotransposition; sex chromosomes.
Figures



Similar articles
-
Adaptive evolution of genes duplicated from the Drosophila pseudoobscura neo-X chromosome.Mol Biol Evol. 2010 Aug;27(8):1963-78. doi: 10.1093/molbev/msq085. Epub 2010 Mar 29. Mol Biol Evol. 2010. PMID: 20351054 Free PMC article.
-
Genes Relocated Between Drosophila Chromosome Arms Evolve Under Relaxed Selective Constraints Relative to Non-Relocated Genes.J Mol Evol. 2018 Jul;86(6):340-352. doi: 10.1007/s00239-018-9849-5. Epub 2018 Jun 21. J Mol Evol. 2018. PMID: 29926120
-
General gene movement off the X chromosome in the Drosophila genus.Genome Res. 2009 May;19(5):897-903. doi: 10.1101/gr.088609.108. Epub 2009 Feb 27. Genome Res. 2009. PMID: 19251740 Free PMC article.
-
Gene content evolution on the X chromosome.Curr Opin Genet Dev. 2008 Dec;18(6):493-8. doi: 10.1016/j.gde.2008.09.006. Epub 2008 Oct 16. Curr Opin Genet Dev. 2008. PMID: 18929654 Free PMC article. Review.
-
Evolution of gene function on the X chromosome versus the autosomes.Genome Dyn. 2007;3:101-118. doi: 10.1159/000107606. Genome Dyn. 2007. PMID: 18753787 Review.
Cited by
-
Little evidence for demasculinization of the Drosophila X chromosome among genes expressed in the male germline.Genome Biol Evol. 2012;4(10):1007-16. doi: 10.1093/gbe/evs077. Epub 2012 Sep 12. Genome Biol Evol. 2012. PMID: 22975718 Free PMC article.
-
Induction and inhibition of Drosophila X chromosome gene expression are both impeded by the dosage compensation complex.G3 (Bethesda). 2022 Aug 25;12(9):jkac165. doi: 10.1093/g3journal/jkac165. G3 (Bethesda). 2022. PMID: 35792851 Free PMC article.
-
Role of testis-specific gene expression in sex-chromosome evolution of Anopheles gambiae.Genetics. 2011 Nov;189(3):1117-20. doi: 10.1534/genetics.111.133157. Epub 2011 Sep 2. Genetics. 2011. PMID: 21890740 Free PMC article.
-
The roles of gene duplications in the dynamics of evolutionary conflicts.Proc Biol Sci. 2024 Jun;291(2024):20240555. doi: 10.1098/rspb.2024.0555. Epub 2024 Jun 12. Proc Biol Sci. 2024. PMID: 38865605 Free PMC article. Review.
-
Comparative Genomic Hybridization (CGH) reveals a neo-X chromosome and biased gene movement in stalk-eyed flies (genus Teleopsis).PLoS Genet. 2010 Sep 16;6(9):e1001121. doi: 10.1371/journal.pgen.1001121. PLoS Genet. 2010. PMID: 20862308 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

