Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution
- PMID: 20333194
- PMCID: PMC2817414
- DOI: 10.1093/gbe/evp019
Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution
Abstract
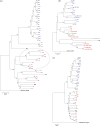
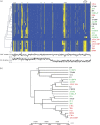
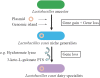
Lactobacillus casei is remarkably adaptable to diverse habitats and widely used in the food industry. To reveal the genomic features that contribute to its broad ecological adaptability and examine the evolution of the species, the genome sequence of L. casei ATCC 334 is analyzed and compared with other sequenced lactobacilli. This analysis reveals that ATCC 334 contains a high number of coding sequences involved in carbohydrate utilization and transcriptional regulation, reflecting its requirement for dealing with diverse environmental conditions. A comparison of the genome sequences of ATCC 334 to L. casei BL23 reveals 12 and 19 genomic islands, respectively. For a broader assessment of the genetic variability within L. casei, gene content of 21 L. casei strains isolated from various habitats (cheeses, n = 7; plant materials, n = 8; and human sources, n = 6) was examined by comparative genome hybridization with an ATCC 334-based microarray. This analysis resulted in identification of 25 hypervariable regions. One of these regions contains an overrepresentation of genes involved in carbohydrate utilization and transcriptional regulation and was thus proposed as a lifestyle adaptation island. Differences in L. casei genome inventory reveal both gene gain and gene decay. Gene gain, via acquisition of genomic islands, likely confers a fitness benefit in specific habitats. Gene decay, that is, loss of unnecessary ancestral traits, is observed in the cheese isolates and likely results in enhanced fitness in the dairy niche. This study gives the first picture of the stable versus variable regions in L. casei and provides valuable insights into evolution, lifestyle adaptation, and metabolic diversity of L. casei.
Keywords: comparative genome hybridization; evolution; niche adaptation.
Figures




References
-
- Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics. 2005;21:3665–3666. - PubMed
-
- Acedo-Felix E, Perez-Martinez G. Significant differences between Lactobacillus casei subsp. casei ATCC 393T and a commonly used plasmid-cured derivative revealed by a polyphasic study. Int J Syst Evol Microbiol. 2003;53:67–75. - PubMed
-
- Axelsson L. Lactic acid bacteria: classification and physiology. In: Salminen S, von Wright A, Ouwehand A, editors. Lactic acid bacteria microbiological and functional aspects. 3rd edition. New York: Marcel Dekker Inc; 2004. pp. 1–66.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

