Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): rearrangement and severe truncation of tRNA genes
- PMID: 20333197
- PMCID: PMC2817422
- DOI: 10.1093/gbe/evp027
Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): rearrangement and severe truncation of tRNA genes
Abstract
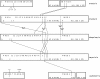
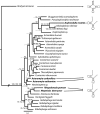
We determined the complete mitochondrial genome sequences of two species of gall midges (Diptera: Cecidomyiidae), as well as partial sequence from a third cecidomyiid and a species from a related family, the Sciaridae. The sciarid sequence has a number of rearrangements of tRNA genes, relative to other dipterans, but is otherwise unremarkable. In contrast, the cecidomyiid genomes possess a number of very unusual features. First, the two complete sequences are very small compared with other dipteran mitochondrial genomes. The genome of Mayetiola destructor is only 14,759 bp while that of Rhopalomyia pomum is only 14,503 bp, comparable with genome sizes observed in some arachnids. Second, all three cecidomyiid species have very high A + T content--more than 83% for the coding region. Third, all three cecidomyiid species possess a number of rearrangements of tRNA genes, including variations within the family. Fourth, the most extraordinary feature of cecidomyiids examined in this study is an extreme truncation of all tRNA genes, including the loss of TPsiC arms and apparent absence of the 3' part of the aminoacyl stems.The truncated tRNA genes of cecidomyiids are very similar to those previously reported for spiders and appear to represent a second, independent origin of these structural features. It is likely that they are made functional through RNA editing, perhaps using the 5' end of the aminoacyl stem as a template for the construction of the required 3' end.
Keywords: Mayetiola destructor; RNA editing; truncated tRNA genes.
Figures

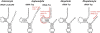
References
-
- Beckenbach AT, Stewart JB. Insect mitochondrial genomics 3: the complete mitochondrial genome sequences of representatives from two neuropteroid orders: a dobsonfly (order Megaloptera), a giant lacewing and an owlfly (order Neuroptera) Genome. 2009;52:31–38. - PubMed
-
- Beuning PJ, Musier-Forsyth K. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymer. 1999;52:1–28. - PubMed
-
- Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. - PubMed

