Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales
- PMID: 20333207
- PMCID: PMC2817432
- DOI: 10.1093/gbe/evp040
Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales
Abstract
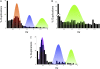
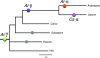
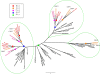
The analysis of the Arabidopsis genome revealed evidence of three ancient polyploidy events in the evolution of the Brassicaceae, but the exact phylogenetic placement of these events is still not resolved. The most recent event is called the At-alpha (alpha) or 3R, the intermediate event is referred to as the At-beta (beta) or 2R, and the oldest is the At-gamma (gamma) or 1R. It has recently been established that At-gamma is shared with other Rosids, including papaya (Carica), poplar (Populus), and grape (Vitis), whereas data to date suggest that At-alpha is Brassicaceae specific. To address more precisely when the At-alpha and At-beta events occurred and which plant lineages share these paleopolyploidizations, we sequenced and analyzed over 4,700 normalized expressed sequence tag sequences from the Cleomaceae, the sister family to the Brassicaceae. Analysis of these Cleome data with homologous sequences from other Rosid genomes (Arabidopsis, Carica, Gossypium, Populus, and Vitis) yielded three major findings: 1) confirmation of a Cleome-specific paleopolyploidization (Cs-alpha) that is independent of the Brassicaceae At-alpha paleopolyploidization; 2) Cleome and Arabidopsis share the At-beta duplication, which is lacking from papaya within the Brassicales; and 3) rates of molecular evolution are faster for the herbaceous annual taxa Arabidopsis and Cleome than the other predominantly woody perennial Rosid lineages. These findings contribute to our understanding of the dynamics of genome duplication and evolution within one of the most comprehensively surveyed clades of plants, the Rosids, and clarify the complex history of the At-alpha, At-beta, and At-gamma duplications of Arabidopsis.
Keywords: Arabidopsis; Brassicales; Cleome; polyploidy; transcriptome.
Figures
References
-
- Ann W, Syring J, Gernandt DS, Liston A, Cronn R. Fossil calibration of molecular divergence infers a moderate mutation rate and recent radiations for Pinus. Mol Biol Evol. 2007;24:90–101. - PubMed
-
- Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. - PubMed

