Pigmentation pathway evolution after whole-genome duplication in fish
- PMID: 20333216
- PMCID: PMC2839281
- DOI: 10.1093/gbe/evp050
Pigmentation pathway evolution after whole-genome duplication in fish
Abstract
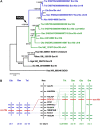
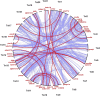
Whole-genome duplications (WGDs) have occurred repeatedly in the vertebrate lineage, but their evolutionary significance for phenotypic evolution remains elusive. Here, we have investigated the impact of the fish-specific genome duplication (FSGD) on the evolution of pigmentation pathways in teleost fishes. Pigmentation and color patterning are among the most diverse traits in teleosts, and their pigmentary system is the most complex of all vertebrate groups. Using a comparative genomic approach including phylogenetic and synteny analyses, the evolution of 128 vertebrate pigmentation genes in five teleost genomes following the FSGD has been reconstructed. We show that pigmentation genes have been preferentially retained in duplicate after the FSGD, so that teleosts have 30% more pigmentation genes compared with tetrapods. This is significantly higher than genome-wide estimates of FSGD gene duplicate retention in teleosts. Large parts of the melanocyte regulatory network have been retained in two copies after the FSGD. Duplicated pigmentation genes follow general evolutionary patterns such as the preservation of protein complex stoichiometries and the overrepresentation of developmental genes among retained duplicates. These results suggest that the FSGD has made an important contribution to the evolution of teleost-specific features of pigmentation, which include novel pigment cell types or the division of existing pigment cell types into distinct subtypes. Furthermore, we have observed species-specific differences in duplicate retention and evolution that might contribute to pigmentary diversity among teleosts.Our study therefore strongly supports the hypothesis that WGDs have promoted the increase of complexity and diversity during vertebrate phenotypic evolution.
Keywords: conserved synteny; fish; functional module; genome duplication; melanocyte; pigment cell.
Figures


References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Aburomia R, Khaner O, Sidow A. Functional evolution in the ancestral lineage of vertebrates or when genomic complexity was wagging its morphological tail. J Struct Funct Genomics. 2003;3:45–52. - PubMed
-
- Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. - PubMed
-
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9:868–882. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

