Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function
- PMID: 20333304
- PMCID: PMC2841643
- DOI: 10.1371/journal.pone.0009773
Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function
Abstract
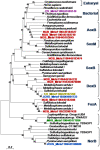
The Yellowstone caldera contains the most numerous and diverse geothermal systems on Earth, yielding an extensive array of unique high-temperature environments that host a variety of deeply-rooted and understudied Archaea, Bacteria and Eukarya. The combination of extreme temperature and chemical conditions encountered in geothermal environments often results in considerably less microbial diversity than other terrestrial habitats and offers a tremendous opportunity for studying the structure and function of indigenous microbial communities and for establishing linkages between putative metabolisms and element cycling. Metagenome sequence (14-15,000 Sanger reads per site) was obtained for five high-temperature (>65 degrees C) chemotrophic microbial communities sampled from geothermal springs (or pools) in Yellowstone National Park (YNP) that exhibit a wide range in geochemistry including pH, dissolved sulfide, dissolved oxygen and ferrous iron. Metagenome data revealed significant differences in the predominant phyla associated with each of these geochemical environments. Novel members of the Sulfolobales are dominant in low pH environments, while other Crenarchaeota including distantly-related Thermoproteales and Desulfurococcales populations dominate in suboxic sulfidic sediments. Several novel archaeal groups are well represented in an acidic (pH 3) Fe-oxyhydroxide mat, where a higher O2 influx is accompanied with an increase in archaeal diversity. The presence or absence of genes and pathways important in S oxidation-reduction, H2-oxidation, and aerobic respiration (terminal oxidation) provide insight regarding the metabolic strategies of indigenous organisms present in geothermal systems. Multiple-pathway and protein-specific functional analysis of metagenome sequence data corroborated results from phylogenetic analyses and clearly demonstrate major differences in metabolic potential across sites. The distribution of functional genes involved in electron transport is consistent with the hypothesis that geochemical parameters (e.g., pH, sulfide, Fe, O2) control microbial community structure and function in YNP geothermal springs.
Conflict of interest statement
Figures


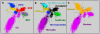
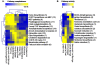
Similar articles
-
Predominant Acidilobus-like populations from geothermal environments in yellowstone national park exhibit similar metabolic potential in different hypoxic microbial communities.Appl Environ Microbiol. 2014 Jan;80(1):294-305. doi: 10.1128/AEM.02860-13. Epub 2013 Oct 25. Appl Environ Microbiol. 2014. PMID: 24162572 Free PMC article.
-
Microbial community structure and sulfur biogeochemistry in mildly-acidic sulfidic geothermal springs in Yellowstone National Park.Geobiology. 2013 Jan;11(1):86-99. doi: 10.1111/gbi.12015. Epub 2012 Nov 21. Geobiology. 2013. PMID: 23231658
-
Linking microbial oxidation of arsenic with detection and phylogenetic analysis of arsenite oxidase genes in diverse geothermal environments.Environ Microbiol. 2009 Feb;11(2):421-31. doi: 10.1111/j.1462-2920.2008.01781.x. Environ Microbiol. 2009. PMID: 19196273
-
Microbes in mercury-enriched geothermal springs in western North America.Sci Total Environ. 2016 Nov 1;569-570:321-331. doi: 10.1016/j.scitotenv.2016.06.080. Epub 2016 Jun 24. Sci Total Environ. 2016. PMID: 27344121 Review.
-
Mining thermophiles for biotechnologically relevant enzymes: evaluating the potential of European and Caucasian hot springs.Extremophiles. 2023 Nov 22;28(1):5. doi: 10.1007/s00792-023-01321-3. Extremophiles. 2023. PMID: 37991546 Free PMC article. Review.
Cited by
-
Microbiome as an endocrine organ and its relationship with eye diseases: Effective factors and new targeted approaches.World J Gastrointest Pathophysiol. 2024 Sep 22;15(5):96446. doi: 10.4291/wjgp.v15.i5.96446. World J Gastrointest Pathophysiol. 2024. PMID: 39355345 Free PMC article. Review.
-
Metagenomics of Thermophiles with a Focus on Discovery of Novel Thermozymes.Front Microbiol. 2016 Sep 27;7:1521. doi: 10.3389/fmicb.2016.01521. eCollection 2016. Front Microbiol. 2016. PMID: 27729905 Free PMC article. Review.
-
Environmental conditions constrain the distribution and diversity of archaeal merA in Yellowstone National Park, Wyoming, U.S.A.Microb Ecol. 2011 Nov;62(4):739-52. doi: 10.1007/s00248-011-9890-z. Epub 2011 Jun 29. Microb Ecol. 2011. PMID: 21713435
-
Geomicrobiology of sublacustrine thermal vents in Yellowstone Lake: geochemical controls on microbial community structure and function.Front Microbiol. 2015 Oct 26;6:1044. doi: 10.3389/fmicb.2015.01044. eCollection 2015. Front Microbiol. 2015. PMID: 26579074 Free PMC article.
-
Phylogenetic and Functional Analysis of Metagenome Sequence from High-Temperature Archaeal Habitats Demonstrate Linkages between Metabolic Potential and Geochemistry.Front Microbiol. 2013 May 15;4:95. doi: 10.3389/fmicb.2013.00095. eCollection 2013. Front Microbiol. 2013. PMID: 23720654 Free PMC article.
References
-
- Streit WR, Schmitz RA. Metagenomics-the key to the uncultured microbes. Curr Opin Microbiol. 2004;7:492–498. - PubMed
-
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, et al. Environmental genome sequencing in the Sargasso Sea. Science. 2004;304:55–74. - PubMed
-
- Tringe S, von Mering GC, Kobayashi A, Salamov AA, Chen K, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

