Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family
- PMID: 20333436
- PMCID: PMC11115768
- DOI: 10.1007/s00018-010-0317-7
Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family
Abstract
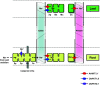
Bacterial Trk and Ktr, fungal Trk and plant HKT form a family of membrane transporters permeable to K(+) and/or Na(+) and characterized by a common structure probably derived from an ancestral K(+) channel subunit. This transporter family, specific of non-animal cells, displays a large diversity in terms of ionic permeability, affinity and energetic coupling (H(+)-K(+) or Na(+)-K(+) symport, K(+) or Na(+) uniport), which might reflect a high need for adaptation in organisms living in fluctuating or dilute environments. Trk/Ktr/HKT transporters are involved in diverse functions, from K(+) or Na(+) uptake to membrane potential control, adaptation to osmotic or salt stress, or Na(+) recirculation from shoots to roots in plants. Structural analyses of bacterial Ktr point to multimeric structures physically interacting with regulatory subunits. Elucidation of Trk/Ktr/HKT protein structures along with characterization of mutated transporters could highlight functional and evolutionary relationships between ion channels and transporters displaying channel-like features.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

