Mouse Rad1 deletion enhances susceptibility for skin tumor development
- PMID: 20334655
- PMCID: PMC2864214
- DOI: 10.1186/1476-4598-9-67
Mouse Rad1 deletion enhances susceptibility for skin tumor development
Abstract
Background: Cells are constantly exposed to stresses from cellular metabolites as well as environmental genotoxins. DNA damage caused by these genotoxins can be efficiently fixed by DNA repair in cooperation with cell cycle checkpoints. Unrepaired DNA lesions can lead to cell death, gene mutation and cancer. The Rad1 protein, evolutionarily conserved from yeast to humans, exists in cells as monomer as well as a component in the 9-1-1 protein complex. Rad1 plays crucial roles in DNA repair and cell cycle checkpoint control, but its contribution to carcinogenesis is unknown.
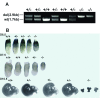
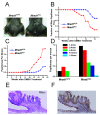
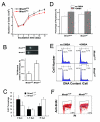
Results: To address this question, we constructed mice with a deletion of Mrad1. Matings between heterozygous Mrad1 mutant mice produced Mrad1+/+ and Mrad1+/- but no Mrad1-/- progeny, suggesting the Mrad1 null is embryonic lethal. Mrad1+/- mice demonstrated no overt abnormalities up to one and half years of age. DMBA-TPA combinational treatment was used to induce tumors on mouse skin. Tumors were larger, more numerous, and appeared earlier on the skin of Mrad1+/- mice compared to Mrad1+/+ animals. Keratinocytes isolated from Mrad1+/- mice had significantly more spontaneous DNA double strand breaks, proliferated slower and had slightly enhanced spontaneous apoptosis than Mrad1+/+ control cells.
Conclusion: These data suggest that Mrad1 is important for preventing tumor development, probably through maintaining genomic integrity. The effects of heterozygous deletion of Mrad1 on proliferation and apoptosis of keratinocytes is different from those resulted from Mrad9 heterozygous deletion (from our previous study), suggesting that Mrad1 also functions independent of Mrad9 besides its role in the Mrad9-Mrad1-Mhus1 complex in mouse cells.
Figures


References
-
- Helt CE, Wang W, Keng PC, Bambara RA. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

