Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294
- PMID: 20335163
- PMCID: PMC2878073
- DOI: 10.1074/jbc.M110.103531
Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294
Abstract
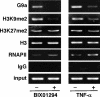
Elucidating the mechanism of human immunodeficiency virus, type 1 (HIV-1) provirus transcriptional silencing in latently infected cells is crucial for understanding the pathophysiological process of HIV-1 infection. It is well established that hypoacetylation of histone proteins by histone deacetylases is involved in the maintenance of HIV-1 latency by repressing viral transcription. Although histone methylation is involved in the organization of chromatin domains and plays a central epigenetic role in gene expression, the role of histone methylation in the maintenance of HIV-1 latency has not been clarified. Here we present evidence that histone H3 Lys(9) (H3K9) methyltransferase G9a is responsible for transcriptional repression of HIV-1 by promoting repressive dimethylation at H3K9 and for the maintenance of viral latency. We observed that G9a significantly inhibited basal, as well as, the induced HIV-1 gene expression by tumor necrosis factor-alpha or Tat. Mutant G9a, however, lacking the SET domain responsible for the catalytic activity of histone methyltransferase, did not show such an effect. When G9a expression was knocked down by small interfering RNA, HIV-1 replication was augmented from cells transiently transfected with a full-length HIV-1 clone. Moreover, a specific inhibitor of G9a, BIX01294, could reactivate expression of HIV-1 from latently infected cells such as ACH-2 and OM10.1. Furthermore, chromatin immunoprecipitation assays revealed the presence of G9a and H3K9 dimethylation on nucleosome histones in the vicinity of the HIV-1 long terminal repeat promoter. These results suggest that G9a is responsible for the transcriptional quiescence of latent HIV-1 provirus and provide a molecular basis for understanding the mechanism by which HIV-1 latency is maintained.
Figures
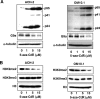
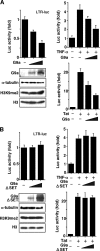
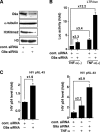
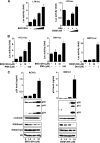

Comment in
-
Reactivation of HIV-1 latent reservoir by an inhibitor of H3K9me2 methyl transferase G9a.Epigenomics. 2010 Aug;2(4):506-7. Epigenomics. 2010. PMID: 22232790 No abstract available.
Similar articles
-
Inhibition of H3K9 histone methyltransferase G9a attenuates renal fibrosis and retains klotho expression.Kidney Int. 2016 Jan;89(1):147-57. doi: 10.1038/ki.2015.291. Epub 2016 Jan 4. Kidney Int. 2016. PMID: 26444031
-
Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation.Virology. 2013 Jun 5;440(2):182-9. doi: 10.1016/j.virol.2013.02.022. Epub 2013 Mar 27. Virology. 2013. PMID: 23541084
-
Multiple Histone Lysine Methyltransferases Are Required for the Establishment and Maintenance of HIV-1 Latency.mBio. 2017 Feb 28;8(1):e00133-17. doi: 10.1128/mBio.00133-17. mBio. 2017. PMID: 28246360 Free PMC article.
-
G9a - An Appealing Antineoplastic Target.Curr Cancer Drug Targets. 2017;17(6):555-568. doi: 10.2174/1568009616666160512145303. Curr Cancer Drug Targets. 2017. PMID: 27174055 Review.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
Cited by
-
Lost in transcription: molecular mechanisms that control HIV latency.Viruses. 2013 Mar 21;5(3):902-27. doi: 10.3390/v5030902. Viruses. 2013. PMID: 23518577 Free PMC article. Review.
-
Mimicking H3 Substrate Arginine in the Design of G9a Lysine Methyltransferase Inhibitors for Cancer Therapy: A Computational Study for Structure-Based Drug Design.ACS Omega. 2021 Feb 22;6(9):6100-6111. doi: 10.1021/acsomega.0c04710. eCollection 2021 Mar 9. ACS Omega. 2021. PMID: 33718701 Free PMC article.
-
Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs.Lancet. 2013 Jun 15;381(9883):2109-17. doi: 10.1016/S0140-6736(13)60104-X. Epub 2013 Mar 29. Lancet. 2013. PMID: 23541541 Free PMC article. Review.
-
Current views on HIV-1 latency, persistence, and cure.Folia Microbiol (Praha). 2017 Jan;62(1):73-87. doi: 10.1007/s12223-016-0474-7. Epub 2016 Oct 5. Folia Microbiol (Praha). 2017. PMID: 27709447 Review.
-
Medicinal Chemistry of Anti-HIV-1 Latency Chemotherapeutics: Biotargets, Binding Modes and Structure-Activity Relationship Investigation.Molecules. 2022 Dec 20;28(1):3. doi: 10.3390/molecules28010003. Molecules. 2022. PMID: 36615199 Free PMC article. Review.
References
-
- Narlikar G. J., Fan H. Y., Kingston R. E. (2002) Cell 108, 475–487 - PubMed
-
- Strahl B. D., Allis C. D. (2000) Nature 403, 41–45 - PubMed
-
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 - PubMed
-
- Berger S. L. (2007) Nature 447, 407–412 - PubMed
-
- Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

