Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication
- PMID: 20335253
- PMCID: PMC2903253
- DOI: 10.1128/JVI.01757-09
Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication
Abstract
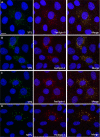
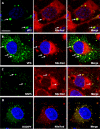
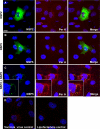
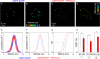
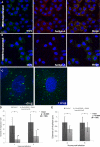
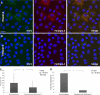
Rotaviruses are a major cause of acute gastroenteritis in children worldwide. Early stages of rotavirus assembly in infected cells occur in viroplasms. Confocal microscopy demonstrated that viroplasms associate with lipids and proteins (perilipin A, ADRP) characteristic of lipid droplets (LDs). LD-associated proteins were also found to colocalize with viroplasms containing a rotaviral NSP5-enhanced green fluorescent protein (EGFP) fusion protein and with viroplasm-like structures in uninfected cells coexpressing viral NSP2 and NSP5. Close spatial proximity of NSP5-EGFP and cellular perilipin A was confirmed by fluorescence resonance energy transfer. Viroplasms appear to recruit LD components during the time course of rotavirus infection. NSP5-specific siRNA blocked association of perilipin A with NSP5 in viroplasms. Viral double-stranded RNA (dsRNA), NSP5, and perilipin A cosedimented in low-density gradient fractions of rotavirus-infected cell extracts. Chemical compounds interfering with LD formation (isoproterenol plus isobutylmethylxanthine; triacsin C) decreased the number of viroplasms and inhibited dsRNA replication and the production of infectious progeny virus; this effect correlated with significant protection of cells from virus-associated cytopathicity. Rotaviruses represent a genus of another virus family utilizing LD components for replication, pointing at novel therapeutic targets for these pathogens.
Figures







Similar articles
-
A Genetically Engineered Rotavirus NSP2 Phosphorylation Mutant Impaired in Viroplasm Formation and Replication Shows an Early Interaction between vNSP2 and Cellular Lipid Droplets.J Virol. 2020 Jul 16;94(15):e00972-20. doi: 10.1128/JVI.00972-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32461314 Free PMC article.
-
Recombinant Rotaviruses Rescued by Reverse Genetics Reveal the Role of NSP5 Hyperphosphorylation in the Assembly of Viral Factories.J Virol. 2019 Dec 12;94(1):e01110-19. doi: 10.1128/JVI.01110-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31619556 Free PMC article.
-
Thiazolides, a new class of antiviral agents effective against rotavirus infection, target viral morphogenesis, inhibiting viroplasm formation.J Virol. 2013 Oct;87(20):11096-106. doi: 10.1128/JVI.01213-13. Epub 2013 Aug 7. J Virol. 2013. PMID: 23926336 Free PMC article.
-
Plasmid-based reverse genetics for probing phosphorylation-dependent viroplasm formation in rotaviruses.Virus Res. 2021 Jan 2;291:198193. doi: 10.1016/j.virusres.2020.198193. Epub 2020 Oct 11. Virus Res. 2021. PMID: 33053412 Free PMC article. Review.
-
Rotavirus replication and the role of cellular lipid droplets: New therapeutic targets?J Formos Med Assoc. 2016 Jun;115(6):389-94. doi: 10.1016/j.jfma.2016.02.004. Epub 2016 Mar 24. J Formos Med Assoc. 2016. PMID: 27017233 Review.
Cited by
-
Peroxisomes and the antiviral responses of mammalian cells.Subcell Biochem. 2013;69:67-75. doi: 10.1007/978-94-007-6889-5_4. Subcell Biochem. 2013. PMID: 23821143 Free PMC article. Review.
-
Lipid Droplet Motility Increases Following Viral Immune Stimulation.Int J Mol Sci. 2021 Apr 23;22(9):4418. doi: 10.3390/ijms22094418. Int J Mol Sci. 2021. PMID: 33922664 Free PMC article.
-
Lactate facilitates classical swine fever virus replication by enhancing cholesterol biosynthesis.iScience. 2022 Oct 13;25(11):105353. doi: 10.1016/j.isci.2022.105353. eCollection 2022 Nov 18. iScience. 2022. PMID: 36339254 Free PMC article.
-
CVB3 Nonstructural 2A Protein Modulates SREBP1a Signaling via the MEK/ERK Pathway.J Virol. 2018 Nov 27;92(24):e01060-18. doi: 10.1128/JVI.01060-18. Print 2018 Dec 15. J Virol. 2018. PMID: 30258014 Free PMC article.
-
Non-Structural Protein-W61 as a Novel Target in Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV): An In-Vitro and In-Silico Study on Protein-Protein Interactions with Nucleoprotein and Viral Replication.Viruses. 2023 Sep 20;15(9):1963. doi: 10.3390/v15091963. Viruses. 2023. PMID: 37766369 Free PMC article.
References
-
- Afrikanova, I., M. C. Miozzo, S. Giambiagi, and O. Burrone. 1996. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 77:2059-2065. - PubMed
-
- Bamford, D. H., and L. Mindich. 2004. Viral molecular machines: replication systems within the inner cores of dsRNA viruses. Virus Res. 101:1.
-
- Bartz, R., J. K. Zehmer, M. Zhu, Y. Chen, G. Serrero, Y. Zhao, and P. Liu. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 6:3256-3265. - PubMed
-
- Becker, W., A. Bergmann, M. A. Hink, K. König, K. Benndorf, and C. Biskup. 2004. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc. Res. Tech. 63:58-66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

