Membrane insertion and biogenesis of the Turnip crinkle virus p9 movement protein
- PMID: 20335263
- PMCID: PMC2876587
- DOI: 10.1128/JVI.00125-10
Membrane insertion and biogenesis of the Turnip crinkle virus p9 movement protein
Abstract
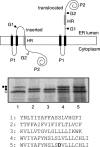
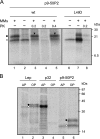
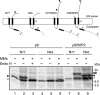
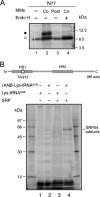
Plant viral infection and spread depends on the successful introduction of a virus into a cell of a compatible host, followed by replication and cell-to-cell transport. The movement proteins (MPs) p8 and p9 of Turnip crinkle virus are required for cell-to-cell movement of the virus. We have examined the membrane association of p9 and found that it is an integral membrane protein with a defined topology in the endoplasmic reticulum (ER) membrane. Furthermore, we have used a site-specific photo-cross-linking strategy to study the membrane integration of the protein at the initial stages of its biosynthetic process. This process is cotranslational and proceeds through the signal recognition particle and the translocon complex.
Figures

Similar articles
-
Double-spanning plant viral movement protein integration into the endoplasmic reticulum membrane is signal recognition particle-dependent, translocon-mediated, and concerted.J Biol Chem. 2005 Jul 8;280(27):25907-12. doi: 10.1074/jbc.M412476200. Epub 2005 May 11. J Biol Chem. 2005. PMID: 15888454
-
Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane.J Biol Chem. 2002 Jun 28;277(26):23447-52. doi: 10.1074/jbc.M202935200. Epub 2002 Apr 25. J Biol Chem. 2002. PMID: 11976343
-
Nuclear localization of turnip crinkle virus movement protein p8.Virology. 2000 Aug 1;273(2):276-85. doi: 10.1006/viro.2000.0440. Virology. 2000. PMID: 10915598
-
Plant virus replication and movement.Virology. 2015 May;479-480:657-71. doi: 10.1016/j.virol.2015.01.025. Epub 2015 Mar 3. Virology. 2015. PMID: 25746797 Review.
-
Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host beta-1,3-glucanases.Semin Cell Dev Biol. 2009 Dec;20(9):1074-81. doi: 10.1016/j.semcdb.2009.05.010. Epub 2009 Jun 6. Semin Cell Dev Biol. 2009. PMID: 19501662 Review.
Cited by
-
Intracellular transport of plant viruses: finding the door out of the cell.Mol Plant. 2011 Sep;4(5):813-31. doi: 10.1093/mp/ssr070. Epub 2011 Sep 5. Mol Plant. 2011. PMID: 21896501 Free PMC article. Review.
-
The Tobacco mosaic virus movement protein associates with but does not integrate into biological membranes.J Virol. 2014 Mar;88(5):3016-26. doi: 10.1128/JVI.03648-13. Epub 2013 Dec 26. J Virol. 2014. PMID: 24371064 Free PMC article.
-
Sequence-dependent scale for translocon-mediated insertion of interfacial helices in membranes.Sci Adv. 2025 Feb 21;11(8):eads6804. doi: 10.1126/sciadv.ads6804. Epub 2025 Feb 19. Sci Adv. 2025. PMID: 39970206 Free PMC article.
-
Membrane integration of poliovirus 2B viroporin.J Virol. 2011 Nov;85(21):11315-24. doi: 10.1128/JVI.05421-11. Epub 2011 Aug 10. J Virol. 2011. PMID: 21835803 Free PMC article.
-
The ER-Membrane Transport System Is Critical for Intercellular Trafficking of the NSm Movement Protein and Tomato Spotted Wilt Tospovirus.PLoS Pathog. 2016 Feb 10;12(2):e1005443. doi: 10.1371/journal.ppat.1005443. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26863622 Free PMC article.
References
-
- Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. - PubMed
-
- Carrington, J. C., L. A. Heaton, D. Zuidema, B. I. Hillman, and T. J. Morris. 1989. The genome structure of turnip crinkle virus. Virology 170:219-226. - PubMed
-
- Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure prediction. Comput. Appl. Biosci. (Camb.) 10:685-686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

