Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs
- PMID: 20335265
- PMCID: PMC2876585
- DOI: 10.1128/JVI.00134-10
Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs
Abstract
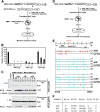
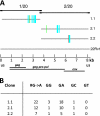
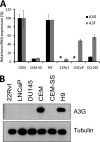
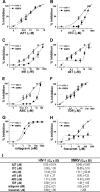
Xenotropic murine leukemia virus-related virus (XMRV), a gammaretrovirus, has been isolated from human prostate cancer tissue and from activated CD4(+) T cells and B cells of patients with chronic fatigue syndrome, suggesting an association between XMRV infection and these two diseases. Since APOBEC3G (A3G) and APOBEC3F (A3F), which are potent inhibitors of murine leukemia virus and Vif-deficient human immunodeficiency virus type 1 (HIV-1), are expressed in human CD4(+) T cells and B cells, we sought to determine how XMRV evades suppression of replication by APOBEC3 proteins. We found that expression of A3G, A3F, or murine A3 in virus-producing cells resulted in their virion incorporation, inhibition of XMRV replication, and G-to-A hypermutation of the viral DNA with all three APOBEC3 proteins. Quantitation of A3G and A3F mRNAs indicated that, compared to the human T-cell lines CEM and H9, prostate cell lines LNCaP and DU145 exhibited 50% lower A3F mRNA levels, whereas A3G expression in 22Rv1, LNCaP, and DU145 cells was nearly undetectable. XMRV proviral genomes in LNCaP and DU145 cells were hypermutated at low frequency with mutation patterns consistent with A3F activity. XMRV proviral genomes were extensively hypermutated upon replication in A3G/A3F-positive T cells (CEM and H9), but not in A3G/A3F-negative cells (CEM-SS). We also observed that XMRV replication was susceptible to the nucleoside reverse transcriptase (RT) inhibitors zidovudine (AZT) and tenofovir and the integrase inhibitor raltegravir. In summary, the establishment of XMRV infection in patients may be dependent on infection of A3G/A3F-deficient cells, and cells expressing low levels of A3G/A3F, such as prostate cancer cells, may be ideal producers of infectious XMRV. Furthermore, the anti-HIV-1 drugs AZT, tenofovir, and raltegravir may be useful for treatment of XMRV infection.
Figures

Similar articles
-
Downregulation of APOBEC3G by xenotropic murine leukemia-virus related virus (XMRV) in prostate cancer cells.Virol J. 2011 Dec 12;8:531. doi: 10.1186/1743-422X-8-531. Virol J. 2011. PMID: 22152111 Free PMC article.
-
Severe restriction of xenotropic murine leukemia virus-related virus replication and spread in cultured human peripheral blood mononuclear cells.J Virol. 2011 May;85(10):4888-97. doi: 10.1128/JVI.00046-11. Epub 2011 Feb 16. J Virol. 2011. PMID: 21325415 Free PMC article.
-
Raltegravir is a potent inhibitor of XMRV, a virus implicated in prostate cancer and chronic fatigue syndrome.PLoS One. 2010 Apr 1;5(4):e9948. doi: 10.1371/journal.pone.0009948. PLoS One. 2010. PMID: 20376347 Free PMC article.
-
The human retrovirus XMRV in prostate cancer and chronic fatigue syndrome.Nat Rev Urol. 2010 Jul;7(7):392-402. doi: 10.1038/nrurol.2010.77. Epub 2010 Jun 1. Nat Rev Urol. 2010. PMID: 20517289 Review.
-
Evidence and controversies on the role of XMRV in prostate cancer and chronic fatigue syndrome.Rev Med Virol. 2011 Jan;21(1):3-17. doi: 10.1002/rmv.673. Epub 2010 Nov 26. Rev Med Virol. 2011. PMID: 21294212 Review.
Cited by
-
Structural and inhibition studies of the RNase H function of xenotropic murine leukemia virus-related virus reverse transcriptase.Antimicrob Agents Chemother. 2012 Apr;56(4):2048-61. doi: 10.1128/AAC.06000-11. Epub 2012 Jan 17. Antimicrob Agents Chemother. 2012. PMID: 22252812 Free PMC article.
-
Discovery of drugs that possess activity against feline leukemia virus.J Gen Virol. 2012 Apr;93(Pt 4):900-905. doi: 10.1099/vir.0.039909-0. Epub 2012 Jan 18. J Gen Virol. 2012. PMID: 22258856 Free PMC article.
-
Downregulation of APOBEC3G by xenotropic murine leukemia-virus related virus (XMRV) in prostate cancer cells.Virol J. 2011 Dec 12;8:531. doi: 10.1186/1743-422X-8-531. Virol J. 2011. PMID: 22152111 Free PMC article.
-
N-linked glycosylation protects gammaretroviruses against deamination by APOBEC3 proteins.J Virol. 2015 Feb;89(4):2342-57. doi: 10.1128/JVI.03330-14. Epub 2014 Dec 10. J Virol. 2015. PMID: 25505062 Free PMC article.
-
Restricted replication of xenotropic murine leukemia virus-related virus in pigtailed macaques.J Virol. 2012 Mar;86(6):3152-66. doi: 10.1128/JVI.06886-11. Epub 2012 Jan 11. J Virol. 2012. PMID: 22238316 Free PMC article.
References
-
- Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317-353. - PubMed
-
- Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

