PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations
- PMID: 20335276
- PMCID: PMC2859132
- DOI: 10.1093/bioinformatics/btq126
PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations
Abstract
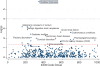
Motivation: Emergence of genetic data coupled to longitudinal electronic medical records (EMRs) offers the possibility of phenome-wide association scans (PheWAS) for disease-gene associations. We propose a novel method to scan phenomic data for genetic associations using International Classification of Disease (ICD9) billing codes, which are available in most EMR systems. We have developed a code translation table to automatically define 776 different disease populations and their controls using prevalent ICD9 codes derived from EMR data. As a proof of concept of this algorithm, we genotyped the first 6005 European-Americans accrued into BioVU, Vanderbilt's DNA biobank, at five single nucleotide polymorphisms (SNPs) with previously reported disease associations: atrial fibrillation, Crohn's disease, carotid artery stenosis, coronary artery disease, multiple sclerosis, systemic lupus erythematosus and rheumatoid arthritis. The PheWAS software generated cases and control populations across all ICD9 code groups for each of these five SNPs, and disease-SNP associations were analyzed. The primary outcome of this study was replication of seven previously known SNP-disease associations for these SNPs.
Results: Four of seven known SNP-disease associations using the PheWAS algorithm were replicated with P-values between 2.8 x 10(-6) and 0.011. The PheWAS algorithm also identified 19 previously unknown statistical associations between these SNPs and diseases at P < 0.01. This study indicates that PheWAS analysis is a feasible method to investigate SNP-disease associations. Further evaluation is needed to determine the validity of these associations and the appropriate statistical thresholds for clinical significance.
Availability: The PheWAS software and code translation table are freely available at http://knowledgemap.mc.vanderbilt.edu/research.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

