Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity
- PMID: 20335359
- PMCID: PMC2853844
- DOI: 10.1242/dev.048678
Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity
Abstract
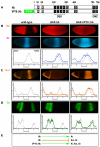
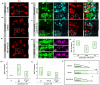
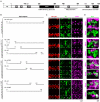
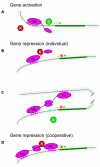
The Hunchback/Ikaros family of zinc-finger transcription factors is essential for specifying the anterior/posterior body axis in insects, the fate of early-born pioneer neurons in Drosophila, and for retinal and immune development in mammals. Hunchback/Ikaros proteins can directly activate or repress target gene transcription during early insect development, but their mode of action during neural development is unknown. Here, we use recombineering to generate a series of Hunchback domain deletion variants and assay their function during neurogenesis in the absence of endogenous Hunchback. Previous studies have shown that Hunchback can specify early-born neuronal identity and maintain 'young' neural progenitor (neuroblast) competence. We identify two conserved domains required for Hunchback-mediated transcriptional repression, and show that transcriptional repression is necessary and sufficient to induce early-born neuronal identity and maintain neuroblast competence. We identify pdm2 as a direct target gene that must be repressed to maintain competence, but show that additional genes must also be repressed. We propose that Hunchback maintains early neuroblast competence by silencing a suite of late-expressed genes.
Figures


References
-
- Albertson R., Chabu C., Sheehan A., Doe C. Q. (2004). Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J. Cell Sci. 117, 6061-6070 - PubMed
-
- Baumgardt M., Karlsson D., Terriente J., Diaz-Benjumea F. J., Thor S. (2009). Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139, 969-982 - PubMed
-
- Berman B. P., Nibu Y., Pfeiffer B. D., Tomancak P., Celniker S. E., Levine M., Rubin G. M., Eisen M. B. (2002). Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99, 757-762 - PMC - PubMed
-
- Berman B. P., Pfeiffer B. D., Laverty T. R., Salzberg S. L., Rubin G. M., Eisen M. B., Celniker S. E. (2004). Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5, R61 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

