Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development
- PMID: 20335407
- PMCID: PMC2852668
- DOI: 10.1093/jxb/erq046
Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development
Abstract
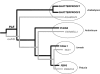
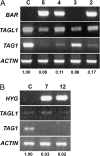
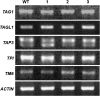
AGAMOUS clade genes encode MADS box transcription factors that have been shown to play critical roles in many aspects of flower and fruit development in angiosperms. Tomato possesses two representatives of this lineage, TOMATO AGAMOUS (TAG1) and TOMATO AGAMOUS-LIKE1 (TAGL1), allowing for an analysis of diversification of function after gene duplication. Using RNAi (RNA interference) silencing, transgenic tomato lines that specifically down-regulate either TAGL1 or TAG1 transcript accumulation have been produced. TAGL1 RNAi lines show no defects in stamen or carpel identity, but show defects in fruit ripening. In contrast TAG1 RNAi lines show defects in stamen and carpel development. In addition TAG1 RNAi lines produce red ripe fruit, although they are defective in determinacy and produce ectopic internal fruit structures. e2814, an EMS- (ethyl methane sulphonate) induced mutation that is temperature sensitive and produces fruit phenotypes similar to that of TAG1 RNAi lines, was also characterized. Neither TAG1 nor TAGL1 expression is disrupted in the e2814 mutant, suggesting that the gene corresponding to the e2814 mutant represents a distinct locus that is likely to be functionally downstream of TAG1 and TAGL1. Based on these analyses, possible modes by which these gene duplicates have diversified in terms of their functions and regulatory roles are discussed.
Figures
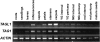



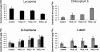
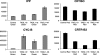

References
-
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

