A pathway-based classification of human breast cancer
- PMID: 20335537
- PMCID: PMC2872436
- DOI: 10.1073/pnas.0912708107
A pathway-based classification of human breast cancer
Abstract
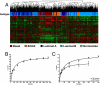
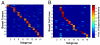
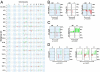
The hallmark of human cancer is heterogeneity, reflecting the complexity and variability of the vast array of somatic mutations acquired during oncogenesis. An ability to dissect this heterogeneity, to identify subgroups that represent common mechanisms of disease, will be critical to understanding the complexities of genetic alterations and to provide a framework to develop rational therapeutic strategies. Here, we describe a classification scheme for human breast cancer making use of patterns of pathway activity to build on previous subtype characterizations using intrinsic gene expression signatures, to provide a functional interpretation of the gene expression data that can be linked to therapeutic options. We show that the identified subgroups provide a robust mechanism for classifying independent samples, identifying tumors that share patterns of pathway activity and exhibit similar clinical and biological properties, including distinct patterns of chromosomal alterations that were not evident in the heterogeneous total population of tumors. We propose that this classification scheme provides a basis for understanding the complex mechanisms of oncogenesis that give rise to these tumors and to identify rational opportunities for combination therapies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Huang E, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. - PubMed
-
- Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

