Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation
- PMID: 20336145
- PMCID: PMC2846535
- DOI: 10.1038/nature08899
Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation
Abstract
Although mammalian hearts show almost no ability to regenerate, there is a growing initiative to determine whether existing cardiomyocytes or progenitor cells can be coaxed into eliciting a regenerative response. In contrast to mammals, several non-mammalian vertebrate species are able to regenerate their hearts, including the zebrafish, which can fully regenerate its heart after amputation of up to 20% of the ventricle. To address directly the source of newly formed cardiomyocytes during zebrafish heart regeneration, we first established a genetic strategy to trace the lineage of cardiomyocytes in the adult fish, on the basis of the Cre/lox system widely used in the mouse. Here we use this system to show that regenerated heart muscle cells are derived from the proliferation of differentiated cardiomyocytes. Furthermore, we show that proliferating cardiomyocytes undergo limited dedifferentiation characterized by the disassembly of their sarcomeric structure, detachment from one another and the expression of regulators of cell-cycle progression. Specifically, we show that the gene product of polo-like kinase 1 (plk1) is an essential component of cardiomyocyte proliferation during heart regeneration. Our data provide the first direct evidence for the source of proliferating cardiomyocytes during zebrafish heart regeneration and indicate that stem or progenitor cells are not significantly involved in this process.
Figures
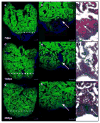
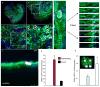
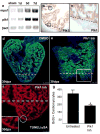
Comment in
-
Thanks be to zebrafish.Circ Res. 2010 Sep 3;107(5):570-2. doi: 10.1161/RES.0b013e3181f6c515. Circ Res. 2010. PMID: 20814025 No abstract available.
References
-
- Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187:249–253. - PubMed
-
- Neff AW, Dent AE, Armstrong JB. Heart development and regeneration in urodeles. Int J Dev Biol. 1996;40:719–725. - PubMed
-
- Flink IL. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat Embryol (Berl) 2002;205:235–244. - PubMed
-
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

