A natural missing link between activated and downhill protein folding scenarios
- PMID: 20336253
- PMCID: PMC7382783
- DOI: 10.1039/b925033f
A natural missing link between activated and downhill protein folding scenarios
Abstract
We propose protein PTB1 : 4W as a good candidate for engineering into a downhill folder. PTB1 : 4W has a probe-dependent thermal unfolding curve and sub-millisecond T-jump relaxation kinetics on more than one time scale. Its refolding rate in denaturant is a non-linear function of denaturant concentration (curved chevron plot). Yet at high denaturant concentration its unfolding is probe-independent, and the folding kinetics can be fitted to a single exponential decay. The domain appears to fold via a mechanism between downhill folding and activated folding over several small barriers, and when denaturant is added, one of these barriers greatly increases and simplifies the observed folding to apparent two-state kinetics. We predict the simplest free energy function consistent with the thermal denaturation and kinetics experiments by using the singular value Smoluchowski dynamics (SVSD) model. PTB1 : 4W is a natural 'missing link' between downhill and activated folding. We suggest mutations that could move the protein into the downhill folding limit.
Figures

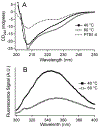
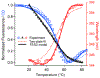
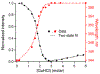
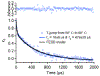


References
-
- Kubelka J; Hofrichter J; Eaton WA. Curr. Opinion. Struct. Bio, 2004, 14, 76–88. - PubMed
-
- Gruebele M. Comptes Rendus Biol, 2005, 328, 701–712. - PubMed
-
- Munoz V. Ann. Rev. Biophys. Biomol. Struct, 2007, 36, 395–412. - PubMed
-
- Zhang J; Li WF; Wang J; Qin M; Wang W. Proteins-Struct. Funct. Bioinformatics, 2008, 72, 1038–1047. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

