Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance
- PMID: 20336341
- PMCID: PMC7088255
- DOI: 10.1007/s10156-010-0045-9
Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance
Abstract
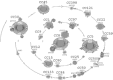
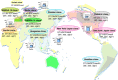
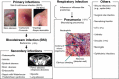
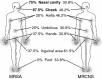
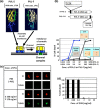
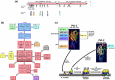
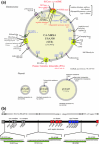
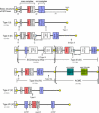
Methicillin-resistant Staphylococcus aureus (MRSA) is able to persist not only in hospitals (with a high level of antimicrobial agent use) but also in the community (with a low level of antimicrobial agent use). The former is called hospital-acquired MRSA (HA-MRSA) and the latter community-acquired MRSA (CA-MRSA). It is believed MRSA clones are generated from S. aureus through insertion of the staphylococcal cassette chromosome mec (SCCmec), and outbreaks occur as they spread. Several worldwide and regional clones have been identified, and their epidemiological, clinical, and genetic characteristics have been described. CA-MRSA is likely able to survive in the community because of suitable SCCmec types (type IV or V), a clone-specific colonization/infection nature, toxin profiles (including Pantone-Valentine leucocidin, PVL), and narrow drug resistance patterns. CA-MRSA infections are generally seen in healthy children or young athletes, with unexpected cases of diseases, and also in elderly inpatients, occasionally surprising clinicians used to HA-MRSA infections. CA-MRSA spreads within families and close-contact groups or even through public transport, demonstrating transmission cores. Re-infection (including multifocal infection) frequently occurs, if the cores are not sought out and properly eradicated. Recently, attention has been given to CA-MRSA (USA300), which originated in the US, and is growing as HA-MRSA and also as a worldwide clone. CA-MRSA infection in influenza season has increasingly been noted as well. MRSA is also found in farm and companion animals, and has occasionally transferred to humans. As such, the epidemiological, clinical, and genetic behavior of CA-MRSA, a growing threat, is focused on in this study.
Figures

Similar articles
-
Molecular characteristics of community-acquired methicillin-resistant Staphylococcus aureus in Hokkaido, northern main island of Japan: identification of sequence types 6 and 59 Panton-Valentine leucocidin-positive community-acquired methicillin-resistant Staphylococcus aureus.Microb Drug Resist. 2011 Jun;17(2):241-50. doi: 10.1089/mdr.2010.0136. Epub 2011 Mar 13. Microb Drug Resist. 2011. PMID: 21395449
-
Genetic diversity of pvl-positive community-onset methicillin-resistant Staphylococcus aureus isolated at a university hospital in Japan.J Infect Chemother. 2017 Dec;23(12):856-858. doi: 10.1016/j.jiac.2017.06.002. Epub 2017 Jun 24. J Infect Chemother. 2017. PMID: 28655502
-
New patterns of methicillin-resistant Staphylococcus aureus (MRSA) clones, community-associated MRSA genotypes behave like healthcare-associated MRSA genotypes within hospitals, Argentina.Int J Med Microbiol. 2014 Nov;304(8):1086-99. doi: 10.1016/j.ijmm.2014.08.002. Epub 2014 Aug 12. Int J Med Microbiol. 2014. PMID: 25240872
-
The evolution of Staphylococcus aureus.Infect Genet Evol. 2008 Dec;8(6):747-63. doi: 10.1016/j.meegid.2008.07.007. Epub 2008 Jul 29. Infect Genet Evol. 2008. PMID: 18718557 Review.
-
Methicillin/Oxacillin-resistant Staphylococcus aureus as a hospital and public health threat in Brazil.Braz J Infect Dis. 2010 Jan-Feb;14(1):71-6. doi: 10.1590/s1413-86702010000100014. Braz J Infect Dis. 2010. PMID: 20428658 Review.
Cited by
-
The epidemiology and molecular characterization of methicillin-resistant staphylococci sampled from a healthy Jordanian population.Epidemiol Infect. 2013 Nov;141(11):2384-91. doi: 10.1017/S0950268813000010. Epub 2013 Jan 23. Epidemiol Infect. 2013. PMID: 23340022 Free PMC article.
-
Stability, Toxicity, and Antibacterial Potential of Gallic Acid-Loaded Graphene Oxide (GAGO) Against Methicillin-Resistant Staphylococcus aureus (MRSA) Strains.Int J Nanomedicine. 2022 Nov 30;17:5781-5807. doi: 10.2147/IJN.S369373. eCollection 2022. Int J Nanomedicine. 2022. PMID: 36474524 Free PMC article.
-
A new local variant (ST764) of the globally disseminated ST5 lineage of hospital-associated methicillin-resistant Staphylococcus aureus (MRSA) carrying the virulence determinants of community-associated MRSA.Antimicrob Agents Chemother. 2013 Apr;57(4):1589-95. doi: 10.1128/AAC.01147-12. Epub 2013 Jan 14. Antimicrob Agents Chemother. 2013. PMID: 23318800 Free PMC article.
-
Pharmacokinetics and pharmacodynamics of aerosolized antibacterial agents in chronically infected cystic fibrosis patients.Clin Microbiol Rev. 2014 Oct;27(4):753-82. doi: 10.1128/CMR.00022-14. Clin Microbiol Rev. 2014. PMID: 25278574 Free PMC article. Review.
-
Evolution and diversity of community-associated methicillin-resistant Staphylococcus aureus in a geographical region.BMC Microbiol. 2011 Sep 29;11:215. doi: 10.1186/1471-2180-11-215. BMC Microbiol. 2011. PMID: 21955438 Free PMC article.
References
-
- Tenover F, Gorwitz RJ. The epidemiology of Staphylococcus infections In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens. 2nd ed. Washington, DC: American Society for Microbiology; 2006. p. 526-34.
-
- Bryskier A. Penicillin. In: Andre B, editor. Antimicrobial agents: antibacterials and antifungals. Washington, DC: American Society for Microbiology; 2005. p. 113–62.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

