Mosquito vitellogenin genes: Comparative sequence analysis, gene duplication, and the role of rare synonymous codon usage in regulating expression
- PMID: 20337554
- PMCID: PMC2999396
- DOI: 10.1673/031.007.0101
Mosquito vitellogenin genes: Comparative sequence analysis, gene duplication, and the role of rare synonymous codon usage in regulating expression
Abstract
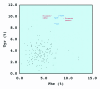
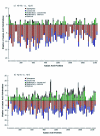
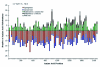
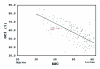
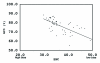
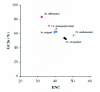
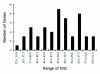
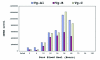
Comparative sequence analysis of mosquito vitellogenin (Vg) genes was carried out to gain a better understanding of their evolution. The genomic clones of vitellogenin genes were isolated and sequenced from all three subfamilies of the family Culicidae including Culicinae (Aedes aegypti, Ochlerotatus atropalpus, Ae. polynesiensis, Ae. albopictus, Ochlerotatus triseriatus and Culex quinquefasciatus), Toxorhynchitinae (Toxorhynchites amboinensis), and Anophelinae (Anopheles albimanus). Genomic clones of vitellogenin genes Vg-B and Vg-C were isolated from Ae. aegypti and sequenced. A comparison of Vg-B and Vg-C, with the previously characterized vitellogenin gene, Vg-A1, suggests that Vg-A1 and Vg-B probably arose by a recent gene duplication, and Vg-C apparently diverged from the two other members of the gene family in an earlier gene duplication event. Two vitellogenin genes orthologous to Vg-C were cloned from a Cx. quinquefasciatus DNA library, one of which is truncated at the N-terminal end. Single vitellogenin genes, orthologous to Vg-C, were cloned from the An. albimanus and Tx. amboinensis libraries. Incomplete sequences orthologous to Vg-B and Vg-C were isolated from the Oc. atropalpus library. Only partial sequences were isolated from Ae. polynesiensis, Ae. albopictus and Oc. triseriatus. Inferred phylogenetic relationships based on analysis of these sequences suggest that Vg-C was the ancestral gene and that a recent gene duplication gave rise to Vg-A1 and Vg-B after the separation of the genus Aedes. The deduced amino acid composition of mosquito vitellogenin proteins exhibits higher tyrosine and phenylalanine composition than other mosquito proteins except for the hexamerin storage proteins. Analysis of vitellogenin coding sequences showed that a majority of amino acid substitutions were due to conserved and moderately conserved changes suggesting that the vitellogenins are under moderately selective constrains to maintain tertiary structure. The vitellogenin genes of the three anautogenous mosquitoes, that require a blood meal to develop eggs, had very high synonymous codon usage biases similar to highly expressed genes of other organisms. On the other hand, the vitellogenin genes of autogenous mosquitoes, that develop at least one batch of eggs without a blood meal, exhibited low synonymous codon usage bias. An unusual pattern of synonymous codon usage was observed in the first 15 amino acid residues encoding the signal peptide in the vitellogenin genes, where a high number of rarely used synonymous codons are present. It is hypothesized that rare synonymous codons have selectively accumulated in the signal peptide region to down-regulate the rate of translation initiation in the absence of a blood meal. Real-time PCR gene expression experiments showed that all three Ae. aegypti vitellogenin genes were highly expressed after a blood meal, and expressed in non-blood-fed females, males, larvae and pupae at trace levels. Sequences were deposited in GenBank (accession numbers: Ae. aegypti Vg-B, AY380797, Vg-C, AY373377; Oc. atropalpus Vg-B, AY691321, Vg-C, AY691322; Ae. polynesiensis Vg-A1, AY691318, Vg-B, AY691319, Vg-C, AY691320; Ae. albopictus Vg-A1, AY691316, Vg-C, AY691317; Oc. triseriatus Vg-C, AY691323; Cx. quinquefasciatus Vg-C1, AY691324, Vg-C2, AY691325; Tx. amboinensis Vg-C, AY691326; An. albimanus Vg-C, AY691327).
Figures
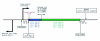


















References
-
- Adamczyk JJ, Fescemyer HW, Heckel DG, Gahan LJ, Davis RE, Kelly TJ. Sex-specific and hormone-controlled expression of a vitellogenin-encoding gene in the gypsy moth. Archives of Insect Biochemistry and Physiology. 1996;31:237–256. - PubMed
-
- Besansky NJ. Codon usage patterns in chromosomal and retrotransposon genes of the mosquito Anopheles gambiae. Insect Molecular Biology. 1993;1:171–178. - PubMed
-
- Burns DM, Beacham IR. Rare codons in Escherichia coli and s-typhimurium signal sequences. FEBS Letters. 1985;189:318–323. - PubMed
-
- Byrne BM, Gruber M, AB G. The evolution of yolk proteins. Progress in Biophysics and Molecular Biology. 1989;53:33–69. - PubMed
-
- Caccone A, Garcia BA, Mathiopoulos KD, Min GS, Moriyama EN, Powell JR. Characterization of the soluble guanylyl cyclase beta-subunit gene in the mosquito Anopheles gambiae. Insect Molecular Biology. 1999;8:23–30. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

